Superbug's Pick'n'Mix DNA
On 18 June 2004, the BBC reported fears that deaths from the ‘superbug’ MRSA (methicillin-resistant Staphylococcus aureus) might double in the next five years. While all reports stress the need for improved cleanliness as a vital preventive measure, medicine is crying out for new ways to tackle MRSA and to monitor its progress.
MRSA is resistant to the majority of antibiotics commonly used in hospitals and there are fears that it might acquire resistance to the remaining effective antibiotic. The number of cases in the UK has grown 600% in eight years.
Today (24 June 2004), researchers from The Wellcome Trust Sanger Institute and their colleagues announce the completion of the genome sequence of an MRSA strain and its antibiotic-sensitive ‘cousin’ MSSA. The strain of MRSA chosen, which causes half of all UK outbreaks, contains several unique genetic elements that are related to its virulence and drug resistance. It has emerged as a major threat in only the past ten years.
“This new atlas shows that the strain that is most prevalent in the UK is very different from previously sequenced superbugs. It has evolved a ‘Pick’n’Mix’ genome that incorporates favoured genes that improve its resistance and ability to cause disease – genes that are vital to its spread and success.”
Dr Matt Holden Leader of the S. aureus analysis team at the Wellcome Trust Sanger Institute
Both strains show evidence of movement of DNA between organisms, acquiring DNA segments from other isolates. In MRSA, some of these new segments may help the organism to thrive in hospitals. More than 6% of its genes are novel.
“MRSA and other pathogens are driven to acquire new tricks to evade or defeat the defences we put up. The dramatic and rapid evolution of S. aureus species underlines the importance of this kind of research to understand how bacteria evolve and how they acquire these new tools.
“Our results clearly show that bacteria are capable of very rapid evolutionary change and remain a formidable enemy.”
Dr Julian Parkhill Project Manager at the Wellcome Trust Sanger Institute
Staphylococcus aureus, the species of bacteria that includes MRSA and MSSA, appear to evolve extremely rapidly and today’s report suggests that improved methods will be required to track S. aureus outbreaks. The genome sequence will help in this.
“The rise in hospital-acquired infections and the increase in resistance to antibiotics are major challenges to health service providers around the world. The genome sequence and the light it sheds on evolution in MRSA gives us one more tool to understand how disease occurs and possibly to find new methods to combat these diseases.”
Dr Sharon Peacock Honorary Consultant in Clinical Microbiology at the John Radcliffe Hospital, Oxford
DNA elements carrying antibiotic resistance and virulence genes are moved between the sensitive and resistant strains. These exchanges dramatically change the virulence of the organism, adding blocks of genes encoding ‘superantigens’ such as those that cause toxic shock or food poisoning as well as making MRSA more resistant to antibiotics.
Genome sequences produced six years ago have already been used to develop new vaccines for bacterial meningitis, currently in trials.
More information
- Over the past 50 years, populations of Staphylococcus aureus have evolved dramatically, in some cases acquiring an armoury of genes that assist it in attaching to host cells, invading host tissues, colonizing the host, evading the immune system, damaging tissues and resisting the effects of antibiotics. Some strains are now resistant to all common antibiotics – penicillin, vancomycin, cephalosporin and methicillin (and its ‘cousin’ flucloxacillin).
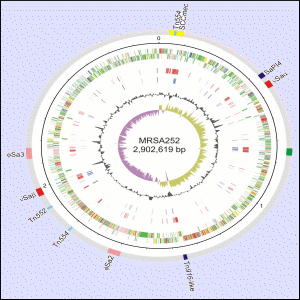
One strain of S. aureus (EMRSA-16) causes one half of all UK MRSA infections and is a major clone found in the US. The sequence generated was from a hospital isolate of the bacterium called MRSA252. The sequence of this was compared with sequence produced for a methicillin-sensitive strain (MSSA476), an isolate that caused severe disease in previously fit child.
The genomes are 2,902,619 base-pairs (2675 genes – MRSA252) and 2,799,802 base-pairs (2566 genes – MSSA476). Although the core order of genes is conserved, comparisons show the genomes are peppered with regions of difference.
- The Nuffield Department of Clinical Medicine is part of the Clinical School in the Medical Sciences Division of the University of Oxford. Biomedical research in Oxford is characterized by its range, depth, multidisciplinary approach and integration. The Department runs one of the largest externally-funded biomedical research programmes in the country, turning over approximately £37m per annum.
Websites
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust and Its Founder
The Wellcome Trust is the most diverse biomedical research charity in the world, spending about £450 million every year both in the UK and internationally to support and promote research that will improve the health of humans and animals. The Trust was established under the will of Sir Henry Wellcome, and is funded from a private endowment, which is managed with long-term stability and growth in mind.


