Periodic table of protein complexes
A new ‘periodic table’ of protein complexes has been developed that provides a unified way to classify and visualise protein complexes, providing a valuable tool for biotechnology and the engineering of novel complexes.
This study also provides insights into evolutionary distribution of different types of existing protein complexes.
The Periodic Table of Protein Complexes, published in Science, offers a new way of looking at the enormous variety of structures that proteins can build in nature, which ones might be discovered next, and predicting how entirely novel structures could be engineered. Created by an interdisciplinary team led by researchers at the Wellcome Genome Campus and the University of Cambridge, the Table provides a valuable tool for research into evolution and protein engineering.
Almost every biological process depends on proteins interacting and assembling into complexes in a specific way, and many diseases are associated with problems in complex assembly. The principles underpinning this organisation are not yet fully understood, but by defining the fundamental steps in the evolution of protein complexes, the new ‘periodic table’ presents a systematic, ordered view on protein assembly, providing a visual tool for understanding biological function.
“Evolution has given rise to a huge variety of protein complexes, and it can seem a bit chaotic. But if you break down the steps proteins take to become complexes, there are some basic rules that can explain almost all of the assemblies people have observed so far.”
Dr Joe Marsh Formerly of the Wellcome Genome Campus and now of the MRC Human Genetics Unit at the University of Edinburgh.
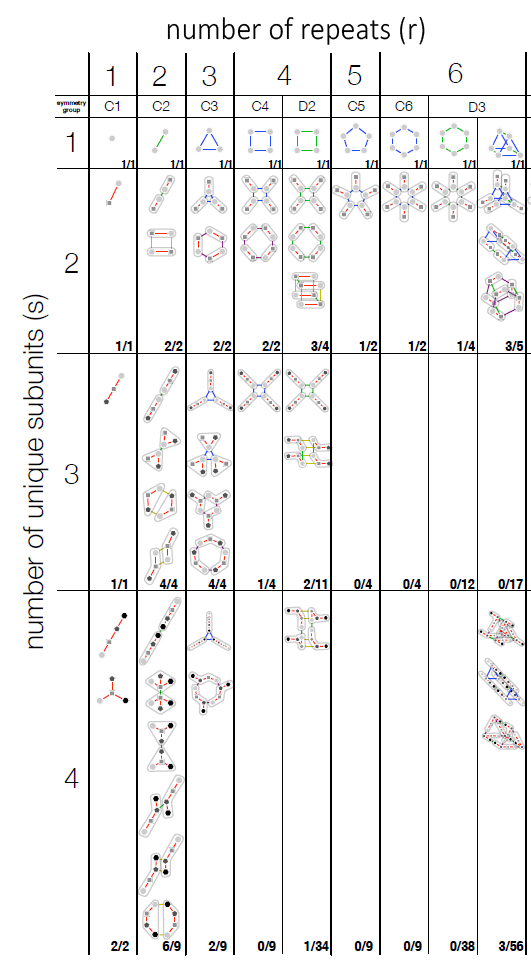
Different ballroom dances can be seen as an endless combination of a small number of basic steps. Similarly, the ‘dance’ of protein complex assembly can be seen as endless variations on dimerization (one doubles, and becomes two), cyclisation (one forms a ring of three or more) and subunit addition (two different proteins bind to each other). Because these happen in a fairly predictable way, it’s not as hard as you might think to predict how a novel protein would form.
“We’re bringing a lot of order into the messy world of protein complexes. Proteins can keep go through several iterations of these simple steps, adding more and more levels of complexity and resulting in a huge variety of structures. What we’ve made is a classification based on these underlying principles that helps people get a handle on the complexity.”
Dr Sebastian Ahnert of the Cavendish Laboratory at the University of Cambridge
The exceptions to the rule are interesting in their own right As are the subject of on-going studies.
“By analysing the tens of thousands of protein complexes for which three-dimensional structures have already been experimentally determined, we could see repeating patterns in the assembly transitions that occur – and with new data from mass spectrometry we could start to see the bigger picture.”
Dr Joe Marsh
“The core work for this study is in theoretical physics and computational biology, but it couldn’t have been done without the mass spectrometry work by our colleagues at Oxford University. This is yet another excellent example of how extremely valuable interdisciplinary research can be.”
Dr Sarah Teichmann Research Group Leader at the Wellcome Trust Sanger Institute and the European Bioinformatics Institute (EMBL-EBI)
More information
Interactive online version of the table
An interactive version of this table with information on the structures represented by each topology can be found at http://www.periodicproteincomplexes.org/
Funding
This work was funded by the Royal Society (S.E.A. and C.V.R.), the Human Frontier Science Program (J.A.M.), the Medical Research Council grant G1000819 (H.H. and C.V.R.) and the Lister Institute for Preventative Medicine (S.A.T.).
Participating Centres
- Theory of Condensed Matter, Cavendish Laboratory, University of Cambridge, JJ Thomson Avenue, Cambridge CB3 0HE
- MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Edinburgh EH4 2XU
- European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD
- Physical and Theoretical Chemistry Laboratory, Department of Chemistry, University of Oxford,South Parks Road, Oxford OX1 3QZ
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA
Publications:
Selected websites
EMBL
EMBL is Europe’s flagship laboratory for the life sciences, with more than 80 independent groups covering the spectrum of molecular biology. EMBL is international, innovative and interdisciplinary – its 1800 employees, from many nations, operate across five sites: the main laboratory in Heidelberg, and outstations in Grenoble; Hamburg; Hinxton, near Cambridge (the European Bioinformatics Institute), and Monterotondo, near Rome. Founded in 1974, EMBL is an inter-governmental organisation funded by public research monies from its member states. The cornerstones of EMBL’s mission are: to perform basic research in molecular biology; to train scientists, students and visitors at all levels; to offer vital services to scientists in the member states; to develop new instruments and methods in the life sciences and actively engage in technology transfer activities, and to integrate European life science research. Around 200 students are enrolled in EMBL’s International PhD programme. Additionally, the Laboratory offers a platform for dialogue with the general public through various science communication activities such as lecture series, visitor programmes and the dissemination of scientific achievements.
EMBL-EBI
The European Bioinformatics Institute is part of EMBL, and is a global leader in the storage, analysis and dissemination of large biological datasets. EMBL-EBI helps scientists realise the potential of ‘big data’ by enhancing their ability to exploit complex information to make discoveries that benefit mankind. We are a non-profit, intergovernmental organisation funded by EMBL’s 21 member states and two associate member states. Our 570 staff hail from 57 countries, and we welcome a regular stream of visiting scientists throughout the year. We are located on the Wellcome Genome Campus in Hinxton, Cambridge in the United Kingdom.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


