Even great mutations need a little help from their friends
Wellcome Trust Sanger Institute scientists have used computer modelling to understand why some mutations in a virus gene rise to dominance and become ‘fixed’ in the genome of the virus, while others die out. Their findings were based upon real-world observations of the evolution of a human flu virus, using genome sequences collected over many years by researchers worldwide.
The team studied mutations in the haemagglutinin gene of the human influenza virus A/H3N2, which latches the virus on to human cells during an infection. Using a computer model, the team identified three key factors that determine the long-term survival of a mutation. The three factors are: the benefit the mutation confers to the virus; the background of other mutations that were present in the gene when the mutation appeared; and the effect of subsequent competition from different versions of the same gene in sibling viruses. To test the influence of each of these factors, they applied their model to the changes seen in the haemagglutinin gene between 1996 and the present day.
“We are keen to understand how evolutionary selection within the flu virus works over time and around the world. In some ways, the flu virus is very simple. When a person catches flu, their cells become incubators for the virus. Within each cell, the virus produces copies of itself. However, the evolution of the virus is not so simple. The copying process is highly error-prone, so that the new viruses often contain mutations. New mutations can either help or hinder the virus from spreading. But having the best mutation doesn’t guarantee evolutionary success.”
“Previous approaches to studying viral evolution have often thought about mutations one at a time, in isolation from each other. But at any one time, a large number of mutations exist. Viruses with different sets of mutations compete against each other, and the combined effect of all of the mutations must be taken into account.”
Dr Chris Illingworth First author of the study, from the Sanger Institute
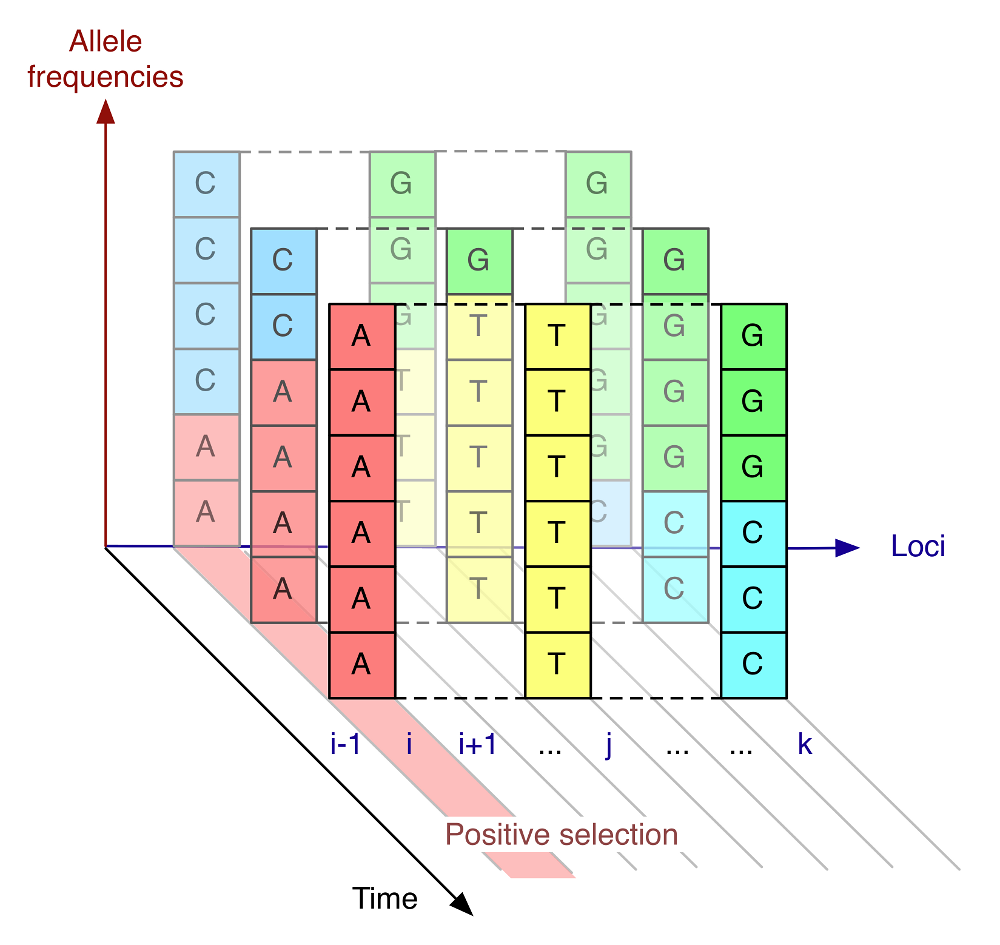
The computer model developed by the researchers considered the benefit, or disadvantage, of each mutation, but also looked at whether pairs of mutations appeared together (in the same virus), or apart (in different viruses). In the model, beneficial mutations in the same virus help each other out, while beneficial mutations in different viruses compete. In the haemagglutinin gene of human influenza A/H3N2 an average of seven new mutations fix in the virus population each year. By analysing which gene variations became dominant and which did not since 1996, the team arrived at their result.
“You might expect that the most beneficial single mutation in a gene would always be the most successful in fixing into the virus’ genome, but this is not what we found. Our analysis of genome sequences of the influenza virus reveals that two other factors are important for a mutation’s success: the sequence of the version of the gene upon which a mutation appeared (how beneficial or not other mutations within the gene are) and the competition from other forms of the gene with different mutations.”
Dr Ville Mustonen Senior author of the paper, from the Sanger Institute
The computer model developed by the team built on methods they had employed for studying evolution in a population using genetic sequence data collected across multiple points in time. The approach accounted for the fact that positive selection – where a beneficial mutation enables a virus to thrive – affects the other mutations occurring across the virus gene, either for their benefit, or for their loss. For example, an unhelpful mutation could succeed if it was coupled to a very beneficial mutation further along the gene – hitchhiking to success. However, a weakly beneficial mutation, if it was opposed by another, stronger mutation, could be caused to die out.
“For the first time, we are able to study the adaptive dynamics of a viral population at the level of individual mutations. We can do this because our method looks at selection across the entire gene, estimating both the relative benefit of the genomic background upon which the mutation appeared, and the combined effect of interference from other mutations on the genome over its lifetime. Statistically, we found that mutations were more likely to become fixed in the flu population if they were beneficial for the virus, had been fortunate to appear on more beneficial backgrounds, and had encountered less opposition from other mutations.”
Chris Illingworth
The new computer model could potentially aid in predicting how the flu virus might change in the future. However Further research is needed to discover the true potential of this approach.
“We need to be cautious before making bold claims. Our mathematical model, like all such approaches, is based on a number of assumptions. The next question to answer is whether or not the model matches reality closely enough to make useful predictions.”
Ville Mustonen
More information
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


