Study doubles number of known human structural variants
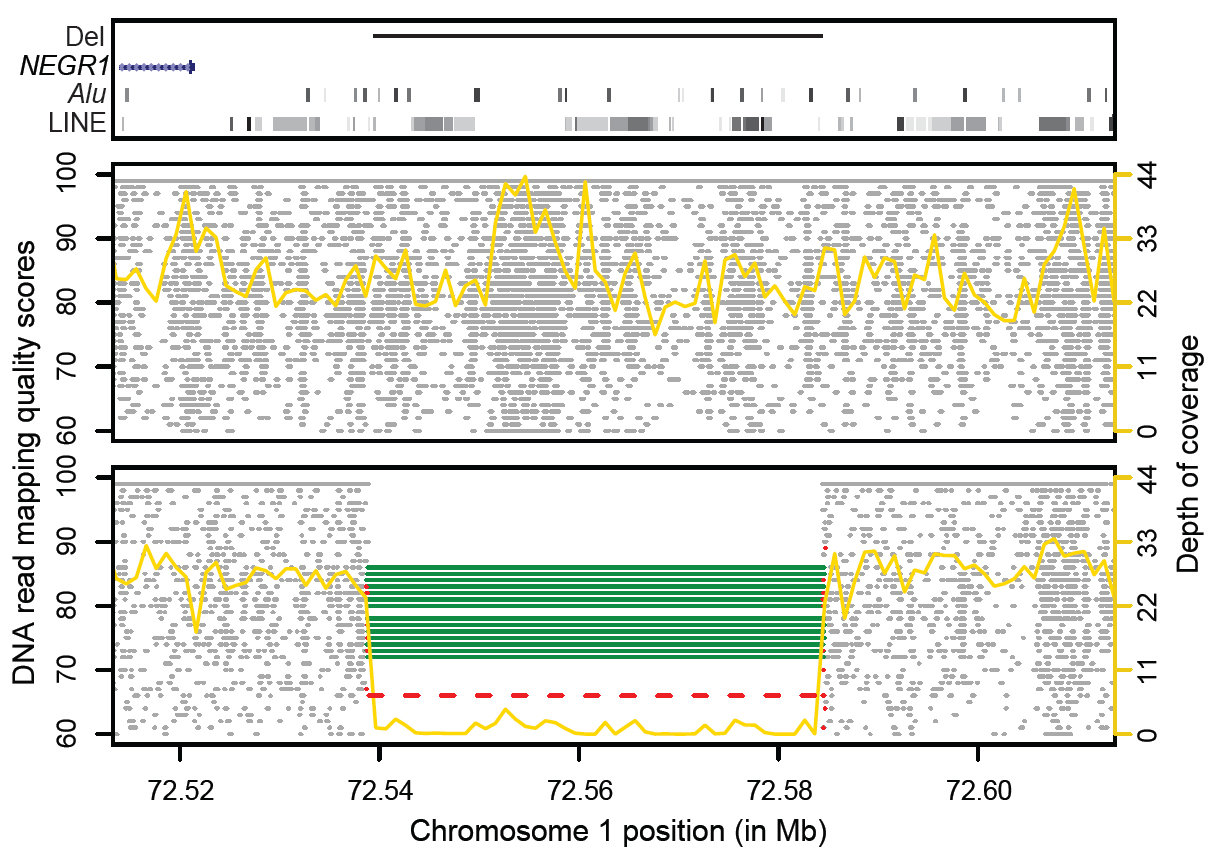
Researchers have created the most detailed map of structural variation in the human genome. The map, which charts approaching 30,000 structural variants – half of them never seen before – will allow teams around the world to look how this kind of genetic effect can shape human health and disease.
Identifying genetic differences between people often involves a search for single letter changes in the genome, which can be associated with disease symptoms or susceptibility to developing a disease. But these single letter changes account for only part of the variability in human genomes. Each person’s genome differs in other ways, carrying an enormous amount of structural variation – deletions, duplications, insertions, and inversions in the genetic sequence. Structural variation is known to play an important role in diseases including autism, schizophrenia and Crohn’s disease.
There are many structural variants in everyone’s genomes and they are increasingly being associated with various aspects of human health. It is important to be able to identify and comprehensively characterise these genetic variants using state-of-the-art DNA sequencing technologies.”
Charles Lee PhD, a clinical cytogeneticist at Brigham and Women’s Hospital and associate professor at Harvard Medical School, and co-chair of this project
The new research goes one step further than previous efforts by using DNA sequencing technology to precisely describe the structural changes – down to the single letter. In the past, finding structural variants has relied on microarray technology, which has been successful at picking out areas of the genome where large structural changes occur. However, researchers were not typically able to scrutinise the detail of the genetic sequences in and around these variants.
The research draws on a pool of 185 genome sequences, produced as part of the pilot phase of the 1000 Genomes Project.
“The data that we need to better appreciate the biomedical consequences of structural variation is now at our fingertips. This research was built upon a highly collaborative international effort, which is creating a fundamental resource for all biomedical scientists. We have now developed a new framework for analysing these key genetic variations that previously received little attention.
“Today’s results provide proof-of-principle – the framework we have developed is now being adopted for other large-scale studies of structural variation.”
Dr Matt Hurles from the Wellcome Trust Sanger Institute and co-chair of the project
The team used their approach to look for smaller structural changes than previously possible – down to 50 DNA letters long. The average size of the variants the team found was around 700 letters long – considerably less than in previous studies. The team also found new structural changes that appear less frequently in humans. The results highlight the power of the researchers’ high-resolution approach to pick out variants that were invisible to previous studies.
As the 1000 Genomes Project scales up from its pilot phase, the researchers will apply their approach to the growing data set, on the way towards a comprehensive catalogue of structural variation found in humans.
The study provided important insights into how structural variants are formed in the genome, thereby linking structural variation to mutational processes acting in the germline.
“We found 51 hotspots where structural variants, such as large deletions, appear to occur particularly often. Six of those hotspots are in regions known to be related to genetic conditions, such as Miller-Dieker syndrome, a congenital brain disease that may lead to infant death.”
Jan Korbel PhD, a senior author of this study from the European Molecular Biology Laboratory in Heidelberg, Germany
As well as finding hotspots associated with genetic disorders, the team found that structural variations that affect certain types of physical process were more common than others. For example, the team discovered that a larger than average amount of their structural variants were involved in cell defence and sensory perception, reinforcing the known role of structural variation in those processes.
The team was also able to make connections between the size of structural variants and the kind of mechanisms by which they were formed.
Together, the results build a profile of the distribution of structural variation – across the genome; across physical processes; and across populations.
“Identifying structural variants from DNA sequencing datasets is very challenging and it is gratifying to see the incredible progress that the structural variation group has made over the past two years.”
Richard Durbin PhD of the Wellcome Trust Sanger Institute and co-chair of the 1000 Genomes Project
“I am confident that this map will serve as an important resource for future sequencing-based disease association studies.”
David Altshuler MD, PhD of the Broad Institute, also a co-chair of the 1000 Genomes Project
Data from the study and ongoing project is being made publically available to the scientific community through the 1000 Genomes Project, an international public-private consortium to build the most detailed map of human genetic variation to date. The 1000 Genomes Project aims to sequence 2500 whole genomes by December 2011, resulting in the largest collection of whole DNA sequencing.
The research was carried out by a consortium of scientists led by Brigham and Women’s Hospital, Harvard Medical School, the Broad Institute, Wellcome Trust Sanger Institute, the University of Washington, and the European Molecular Biology Laboratories in Germany.
More information
Funding
A full list of funding agencies is available at the Nature website.
Participating Centres
A full list of participating centres is available at the Nature website.
Publications:
Selected websites
1000 Genomes Project
Brigham and Women's Hospital
Brigham and Women’s Hospital (BWH) is a 793-bed nonprofit teaching affiliate of Harvard Medical School and a founding member of Partners HealthCare, an integrated health care delivery network. BWH is the home of the Carl J. and Ruth Shapiro Cardiovascular Center, the most advanced center of its kind. BWH is committed to excellence in patient care with expertise in virtually every specialty of medicine and surgery. The BWH medical preeminence dates back to 1832, and today that rich history in clinical care is coupled with its national leadership in quality improvement and patient safety initiatives and its dedication to educating and training the next generation of health care professionals. Through investigation and discovery conducted at its Biomedical Research Institute (BRI), BWH is an international leader in basic, clinical and translational research on human diseases, involving more than 900 physician-investigators and renowned biomedical scientists and faculty supported by more than $ 537 M in funding. BWH is also home to major landmark epidemiologic population studies, including the Nurses’ and Physicians’ Health Studies and the Women’s Health Initiative.
The Eli and Edythe L. Broad Institute of MIT and Harvard
The Eli and Edythe L. Broad Institute of MIT and Harvard, founded in 2003 by MIT, Harvard and its affiliated hospitals, and Los Angeles philanthropists Eli and Edythe L. Broad, includes faculty, professional staff and students from throughout the MIT and Harvard biomedical research communities and beyond, with collaborations spanning over a hundred private and public institutions in more than 40 countries worldwide.
The European Molecular Biology Laboratory
The European Molecular Biology Laboratory is a basic research institute funded by public research monies from 20 member states (Austria, Belgium, Croatia, Denmark, Finland, France, Germany, Greece, Iceland, Ireland, Israel, Italy, Luxembourg, the Netherlands, Norway, Portugal, Spain, Sweden, Switzerland and the United Kingdom) and associate member state Australia. Research at EMBL is conducted by approximately 85 independent groups covering the spectrum of molecular biology. The Laboratory has five units: the main Laboratory in Heidelberg, and Outstations in Hinxton (the European Bioinformatics Institute), Grenoble, Hamburg, and Monterotondo near Rome. The cornerstones of EMBL’s mission are: to perform basic research in molecular biology; to train scientists, students and visitors at all levels; to offer vital services to scientists in the member states; to develop new instruments and methods in the life sciences and to actively engage in technology transfer activities. Around 190 students are enrolled in EMBL’s International PhD programme. Additionally, the Laboratory offers a platform for dialogue with the general public through various science communication activities such as lecture series, visitor programmes and the dissemination of scientific achievements.
NHGRI
NHGRI is one of 27 institutes and centers at the NIH, an agency of the Department of Health and Human Services. The NHGRI Division of Extramural Research supports grants for research and for training and career development sites nationwide.
The National Institutes of Health
The National Institutes of Health – “The Nation’s Medical Research Agency” – includes 27 institutes and centers, and is a component of the U.S. Department of Health and Human Services. It is the primary U.S. federal agency for conducting and supporting basic, clinical and translational medical research, and it investigates the causes, treatments and cures for both common and rare diseases.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


