Candida can do less
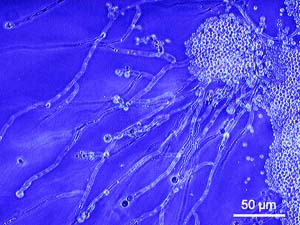
Sanger Institute researchers and their collaborators have published the genome of a species of Candida that is closely related to the most pathogenic of human yeasts, Candida albicans. The new sequence of the less virulent relative, Candida dubliniensis, suggests that it has lost many genes found in C. albicans and thereby diminishing its potency as a pathogen. The work was a collaboration from labs in the UK, Ireland, Austria and New Zealand.
Candida species are pathogenic yeasts that cause superficial infections of the mouth or vagina (commonly called thrush) as well as deeper infections that can be life threatening. In most cases, the cause of the infection is the C. albicans species. As the closest relative of C. albicans, C. dubliniensis shares the vast majority of its physical characteristics. However, despite striking similarities, C. dubliniensis causes dramatically fewer infections in humans.
“C. albicans and C. dubliniensis are virtually identical in every respect, except that C. dubliniensis very rarely causes disease in humans with healthy immune systems. So we sequenced the C. dubliniensis species and compared it to the available reference genome sequence for C. albicans in the hope that we might be able to associate any genomic differences between the species with the capacity to cause disease.
“The biological differences between these species meant that this comparison was essentially a ‘natural experiment’.”
Dr Andrew Jackson From the Sanger Institute and first author of the study
With good reference genome sequences in place for a variety of disease-causing pathogens like Candida albicans, researchers can begin to engage in comparative genomics. Understanding differences in the genome sequences of related pathogens can, in turn, lead to understanding differences in how they cause disease.
The team found that the two genome sequences were very similar. However, they identified 168 genes specific to each species of yeast. The results suggest that C. albicans has gained potency through the expansion of several gene families. The team identified two gene families in particular that were previously relatively unknown and which appear to confer a distinct advantage on C. albicans in terms of its ability to cause human disease.
The team also found 115 genes that had been inactivated in the C. dubliniensis genome.
“How two species as closely related as C. albicans and C. dubliniensis differ so much in their ability to cause disease has long been a quandary. The presence of expanded novel gene families in C. albicans suggests that there are previously uncharacterised genes involved in candidal virulence, the study of which will enhance our understanding of Candida infections.”
Dr Derek Sullivan From Trinity College Dublin
Dr Sullivan was part of the team that originally discovered Candida dubliniensis and first proposed the genome project.
The team show that while C. albicans and C. dubliniensis have a shared ancestor, their recent evolutionary histories have been remarkably divergent. C. albicans has expanded and diversified important gene families associated with its capacity to cause disease in humans, meanwhile quite the opposite effect can be seen in C. dubliniensis, in fact the species appears to have contracted its genetic repertoire and thereby its disease-causing armoury.
“Comparing these two genome sequences has given us insights into the genetic events and disparities that have led to increased potency in C. albicans. Because the two species are so closely related, we were able to look at a much more recent chapter in the evolution of C. albicans than has been possible in previous Candida genome comparisons.
“The results provide substantial new leads that will allow the research community to take further steps towards characterising the molecular basis of candidal virulence.”
Dr Matt Berriman Leader of eukaryotic pathogen sequencing at the Sanger Institute and senior author on the study
More information
Funding
This work was supported by the Wellcome Trust and grants from Science Foundation Ireland, the European Union and the Irish Health Research Board.
Participating Centres
- Pathogen Genomics Group, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK
- Microbiology Research Unit, Division of Oral Biosciences, Dublin Dental
- School and Hospital, University of Dublin, Trinity College Dublin, Dublin, Ireland
- School of Biomolecular and Biomedical Science, Conway Institute, University
- College Belfield, Dublin, Ireland
- Swammerdam Institute for Life Sciences, University of Amsterdam, The Netherlands. Department of Biochemistry, University of Otago, Dunedin, New Zealand
- University of Aberdeen, Institute of Medical Sciences, Aberdeen, UK
- Medical Statistics and Informatics, Medical University of Vienna, Vienna, Austria
Publications:
Selected websites
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute, which receives the majority of its funding from the Wellcome Trust, was founded in 1992. The Institute is responsible for the completion of the sequence of approximately one-third of the human genome as well as genomes of model organisms and more than 90 pathogen genomes. In October 2006, new funding was awarded by the Wellcome Trust to exploit the wealth of genome data now available to answer important questions about health and disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


