ONE STEP tagging
Technology
The ability to engineer the human genome is extremely important in fundamental research, disease modelling and in the development of cellular therapeutics. CRISPR-Cas9, piggyBac and Lentivirus are powerful tools that have been used to generate defined mutations and integrate transgenes at specific sites in the genome, but with several limitation, especially for the introduction of large exogenous sequences. To circumvent some of these problems, we have developed, and patented, a technique that allows simple and efficient integration of large transgenes at essentially any site within the genome using a combination of CRISPR/Cas9 and site-specific recombination, defined as ONE STEP tagging.
A novel gene editing-method that allows for integration of large transgenes at specific sites in the genome using a combination of CRISPR-Cas9 and site-specific recombination in a single step defined as ONE STEP tagging.
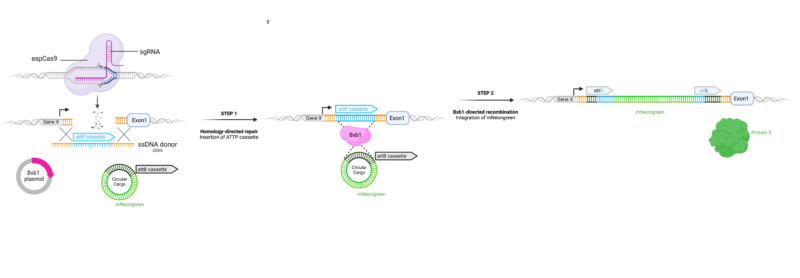
- ONE STEP tagging is achieved through the simultaneous delivery of:
- Cas endonuclease ribonucleoprotein (RNP) complex
- Short single stranded oligodeoxynucleotide (ssODN) containing a recombination cassette (attP)
- Plasmid expressing a Bxb1 recombinase
- Plasmid containing the universal donor of choice and a recombination cassette (attB)
- The Cas endonuclease RNP complex generates a site-specific cut in the genome which upon repair can utilise the ssODN as HDR template, resulting in introduction of a recombinase cassette (attP) site into a specific genomic locus.
- Upon expression, the Bxb1 recombinase recognises the integrated, now double-stranded recombination cassette in the genomic locus (attP) and the universal donor plasmid (attB) and facilitates the integration of the donor at the defined locus.
Advantages
- Precise genomic integration of transgenes at any endogenous loci through a single transfection event.
- Integration of large transgenes, otherwise challenging to achieve via CRISPR-mediated HDR.
- Itilises universal cargo designs, allowing for targeting of different sites in a highly modular way while by-passing tedious processing steps.
- ONE STEP tagging cargos are versatile and cover a wide range of tagging options e.g. fluorescent, degron or purification tags or other specific cargos (e.g. CARs)
Applications/Context
ONE STEP tagging has the potential for application in various research and development and translational settings.
R&D setting
Genes tagged by the ONE STEP technology are suitable for a number of applications:
- Lineage tracing experiments aim to delineate all the progeny produced by a single cell or a group of cells.
- Analysis of protein dynamics and localisation.
- Immunoprecipitation of endogenously tagged proteins where appropriate antibodies are currently unavailable.
- Screening of transcription factors or endogenous promoter dynamics and characterisation.
- Potential of simultaneous tagging of multiple loci through the use of heterotypic cassettes and a variety of donors (see figure below).
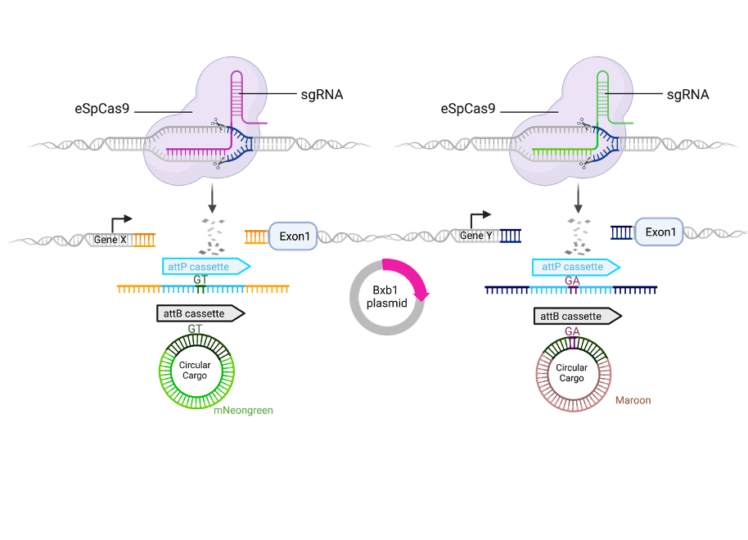
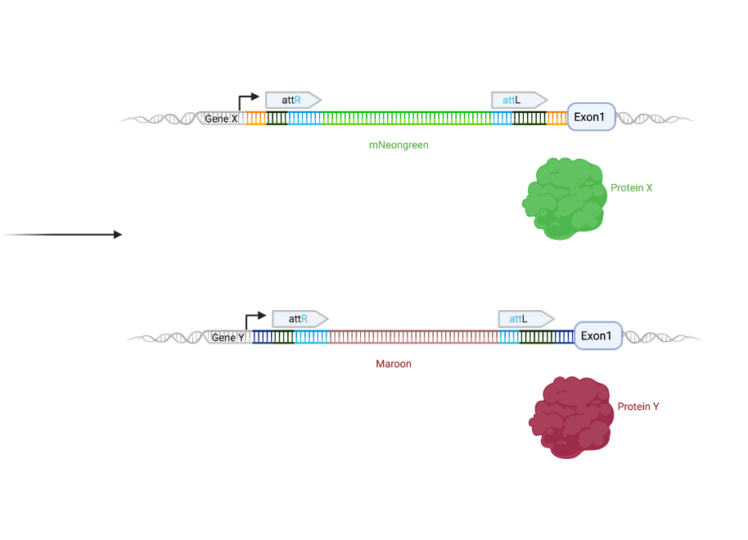
Translational setting
Potential clinical applications:
- Integration of exogenous promoter driven transcription factors to drive lineage specific differentiation from iPSCs (i.e. HSCs, neuronal lines, etc.)
- Generation of ‘off-the-shelf’ CART cells resistant to alloresponsive T -cell attacks and with reduced exhaustion. This could be achieved by combining ONE STEP tagging at the TCR locus with targeted knock-out at the B2M and PD-1 loci.
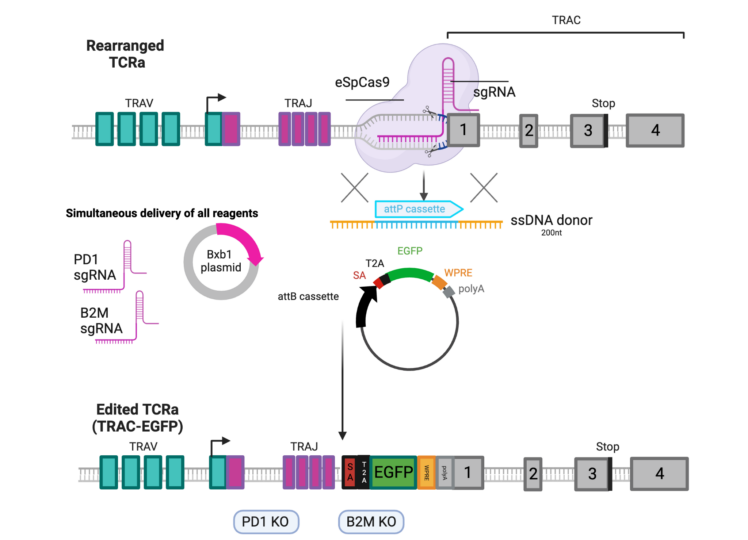
Comparable Technologies
ONE STEP tagging technology provides several advantages over other methodologies. mRNA-based approaches are efficient for overexpression of transgenes, however in a transient manner which doesn’t support long-term overexpression of transgenes.
CRISPR-Cas9 is a powerful tool that has been used to generate defined mutations and integrate transgenes at specific genomic sites. However, there are several limitations to this method:
- Generation of large homology directed repair (HDR) constructs for each specific integration is tedious and time consuming to assemble.
- DNA repair in mammalian cells is generally biased against HDR, resulting in undesirable repair outcomes.
- The efficiency of integration via HDR decreases dramatically as the length of the cargo increases, making it challenging to integrate large transgenes.
The use of lentivirus systems for delivery of the genetic material has been shown to be highly prone to transgene silencing (typically 70-80% silenced), resulting in variable expression levels, difficulty in robust production of viral supernatant, and is challenging to regulate the number of integrations per cell. In addition to silencing, the non-specific nature of the integration event(s) means that there is a risk of oncogenic transformation.
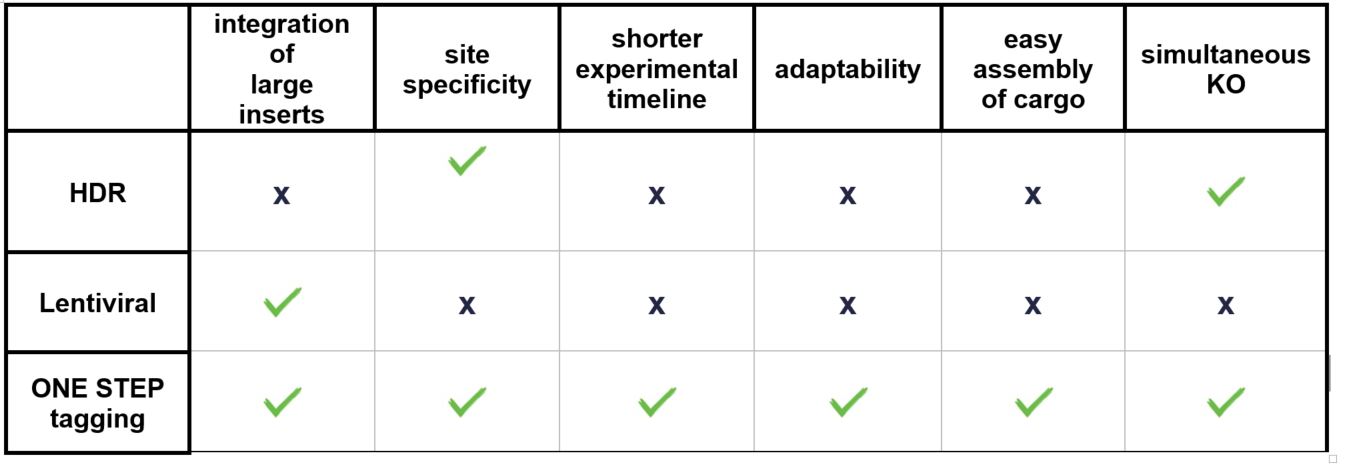
*twin prime editing is a promising technology that does not rely on the generation of double-strand breaks in the genome, but relies on delivery of a larger prime editor enzyme that has been shown to have greater toxicity and reduced efficiency in certain cell types (human pluripotent stem cells) than the Cas9 endonuclease system. Furthermore, identification of suitable target sites is more challenging compared to gRNA based methods.
Intellectual property
Priority patent application filed. The Wellcome Sanger Institute is offering non-exclusive licenses to its IPR.
