Gubbins
Gubbins (Genealogies Unbiased By recomBinations In Nucleotide Sequences) can be used to identify recombination.
Gubbins (Genealogies Unbiased By recomBinations In Nucleotide Sequences) can be used to identify recombination using an algorithm that iteratively identifies loci containing elevated densities of base substitutions while concurrently constructing a phylogeny based on the putative point mutations outside of these regions.
Archive Page
This page is maintained as a historical record and is no longer being updated.
This page was archived in 2022
About
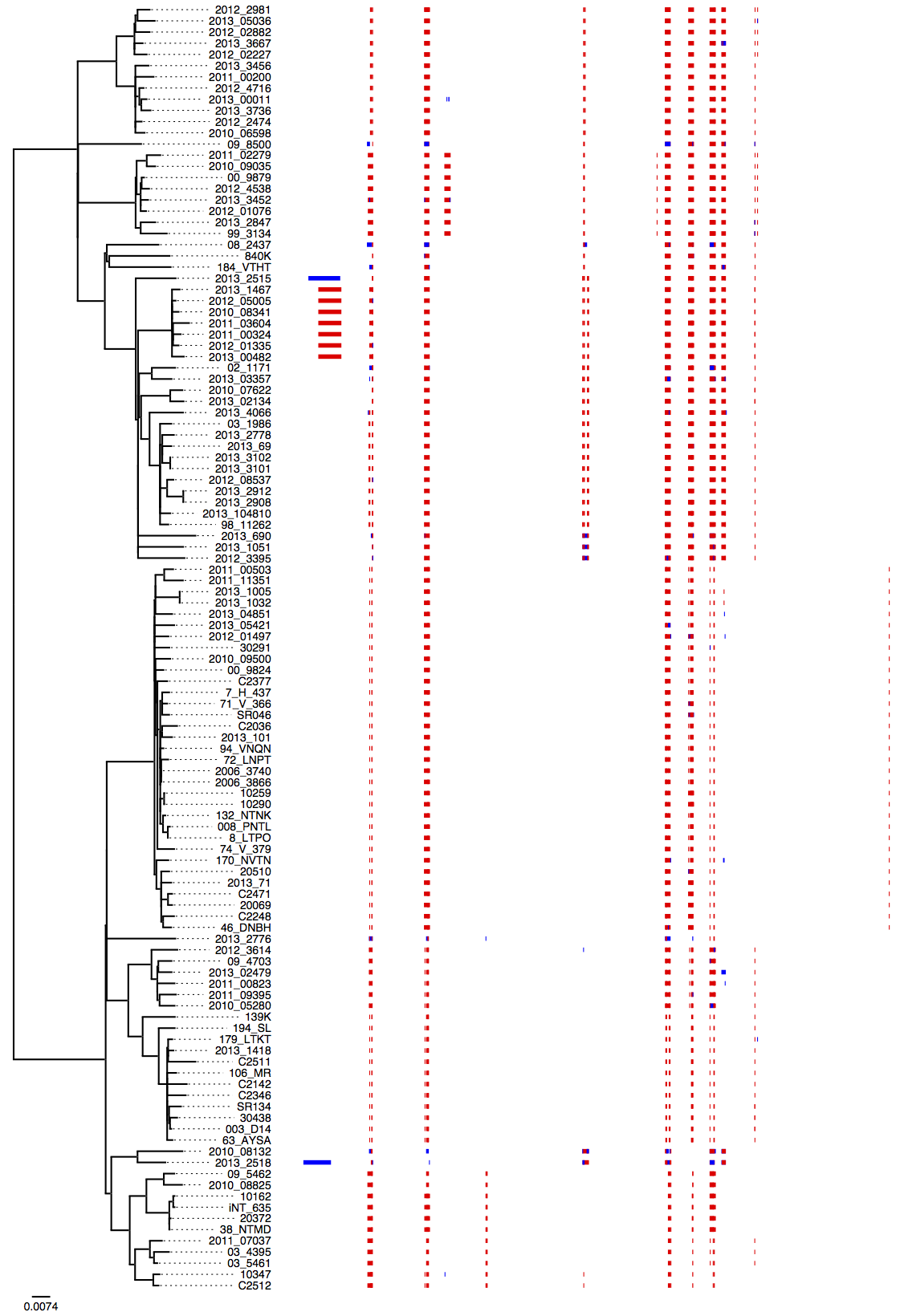
-
Downloads
Please see our GitHub page for download and installation instructions.
Further information
For more information and advice on using this software please see the user manual and our GitHub page.
Gubbins is available under GPL2.
If you make use of this software in your research, please cite as:
Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J and Harris SR
Nucleic acids research 2015;43;3;e15
PUBMED: 25414349; PMC: 4330336; DOI: 10.1093/nar/gku1196
Contact
If you need help or have any queries, please contact us using the details below.
If you need help or have any queries, please contact us using the details below.
- Email: gubbins@sanger.ac.uk
Our normal office hours are 8:30-17:00 (GMT) Monday to Friday.
Sanger Institute Contributors
Previous contributors

Dr Simon Harris
Senior Staff Scientist

Dr Andrew Page
Principal Computer Programmer
