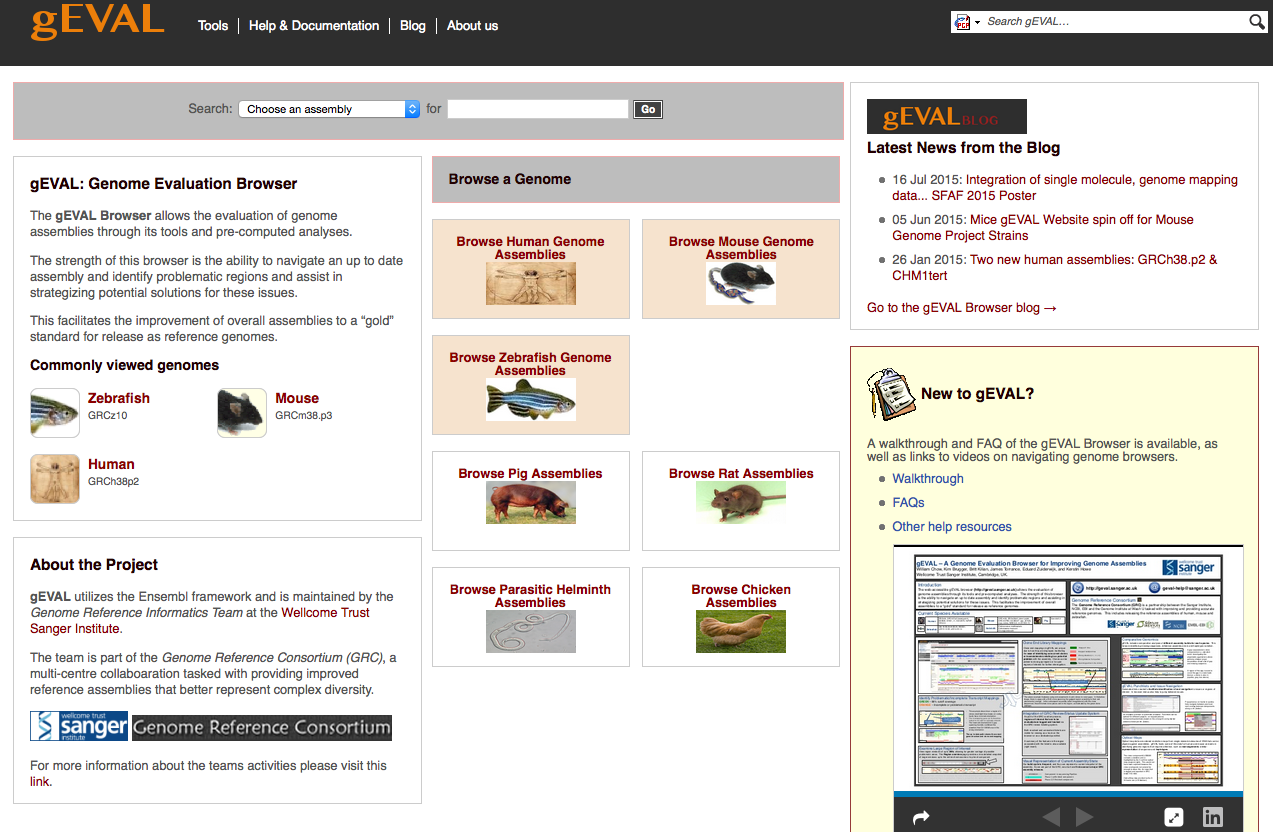
gEVAL - Genome Evaluation Browser
Genome Assembly Evaluation Browser
The gEVAL Browser allows the evaluation of genome assemblies through its tools and pre-computed analyses.
The strength of this browser is the ability to navigate an up to date assembly and identify problematic regions and assist in strategizing potential solutions for these issues.
This facilitates the improvement of overall assemblies to a “gold” standard for release as reference genomes.
Highlights:
- Assemblies for GRC Species (human, mouse, zebrafish) not available in any other public browser.
- Comparative aligments between assemblies within species to aid in gap sizing/closure, confirmation of correct components on path and variation capture.
- Optical and Genome mapping alignments (Opgen, Bionano).
- Lists and navigation tools to problematic regions or regions of interest to aid in correction and finishing.
- Data/analysis tracks are visually designed to aid in quickly identifying regions of interest/problems in the assembly.
About
Downloads
gEVAL can be accessed here:
http://geval.org.uk
The availability of the website plugin code to the public is currently being prepared. Keep an eye out in the space below.
http://wchow.github.io/wtsi-geval-plugin/
Further information
A general walkthrough can be found here:
Contact
If you need help or have any queries, please contact us using the details below.
Sanger Institute Contributors

Dr Kerstin Howe
Head of Production Genomics
Previous contributors

William Chow
Senior Bioinformatician