Teichmann Group
Gene expression genomics
Sarah Teichmann is co-founder and principal leader of the Human Cell Atlas (HCA) international consortium. The International Human Cell Atlas initiative aims to create comprehensive reference maps of all human cells to further understand health and disease.
The team’s research focuses on four complementary study areas:
- Transcriptional regulation and gene expression
- Single-cell genomics
- Immunology
- Protein complexes
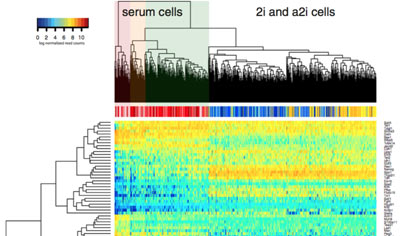 Transcriptional regulation and gene expression
Transcriptional regulation and gene expression
We study the processes that control cell-specific gene expression in both embryonic stem cells and in T lymphocytes.
This research enables us to ask questions about the mechanisms that maintain stem-cell pluripotency and the ways that cells make fate decisions within the immune system.
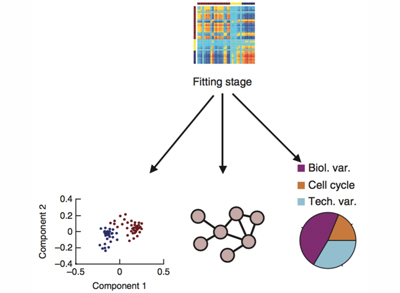 Single-cell genomics
Single-cell genomics
It is clear that differences between cells within seemingly homogenous populations can be crucial in many biological processes.
We work with cutting-edge techniques that enable us to study cell populations at single-cell resolution.
We apply these techniques to better understand our areas of interest and we also develop novel computational approaches to analyse single-cell data.
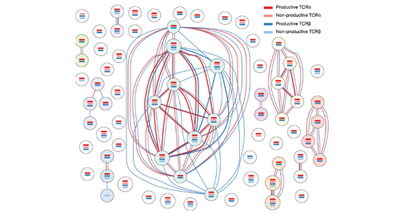 Immunology
Immunology
We are fascinated by the mechanisms that control the development, differentiation and fate choices of CD4+ T lymphocytes.
We study these cells at the single-cell level to understand their behaviour during immune responses and also the role of the cells’ specific T cell receptors.
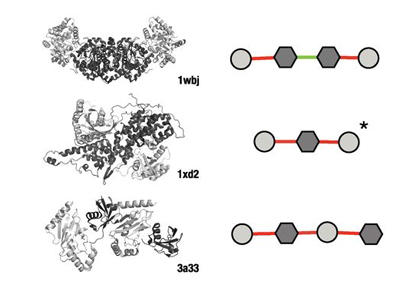 Protein complexes
Protein complexes
We investigate the principles that govern the folding and assembly of protein complexes. Most recently, by analysing the tens of thousands of protein complexes for which three-dimensional structures have already been experimentally determined, we found repeating patterns in the assembly transitions that occur, which could be organized into a periodic table.
20th Anniversary Celebration

Seminars from the 20th Anniversary Celebration
Aleksandra Ola Kolodziejczyk
Emmanuel Levy
Joe Marsh
Jose Leal
Kerstin Meyer
M Madan Babu
Roser Vento
Tina Perica
Tzachi Hagai
Valentine Svensson
Core team

Dr Benjamin Stewart
Postdoctoral Researcher

Chenqu Suo
PhD Student
Previous core team members

Tracey Andrew
Human Cell Atlas Operations Manager

Dr Emmanouil Athanasiadis
Visiting Worker - Research Associate

Adrian Bird
Former Associate Faculty

Xi Chen
Senior Staff Scientist

Dr Cecilia Domínguez Conde
Postdoctoral Fellow

Tomas Pires de Carvalho Gomes
PhD Student

Dr Angela Goncalves
Former Postdoctoral Fellow at the Sanger Institute

Dr Peng He
Visiting Scientist

Dr Kylie James
Postdoctoral Fellow

Dr Vitalii Kleshchevnikov
Bioinformatician

Dr Natsuhiko Kumasaka
Staff Scientist

Monika Litvinukova
Visiting Scientist

Angela Macharia
Project Manager

Elo Madissoon
Postdoctoral Fellow

Lira Mamanova
Scientific Manager

Dr Kerstin Meyer
Principal Staff Scientist

Dr Daniele Muraro
Senior Bioinformatician

Dr Kedar Natarajan
Postdoctoral Fellow

Jongeun Park
Postdoctoral Fellow

Raghd Rostom
PhD Student

Dr Waradon SUNGNAK
Postdoctoral Fellow

Dr Michael Stubbington
Principal Staff Scientist

Dr Carlos Talavera-Lopez
Computational Biologist

Dr Kelvin Tuong
Visiting Scientist
Related groups
Programmes and Facilities
Partners
We work with the following groups
External
Wellcome Trust
External
Chan Zuckerberg Initiative
External
BHF-DZHK
External
Medical Research Council
External
