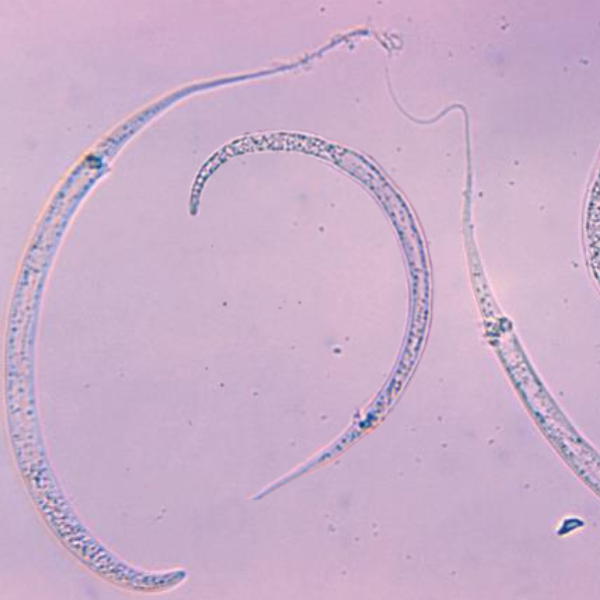
Genomic epidemiology to support Guinea worm eradication
In collaboration with the Guinea worm eradication programme, the parasite genomics group is using genome analysis to determine the epidemiological link between the growing number of canine cases and the persistence of "sporadic" human cases.
The global campaign to eradicate Guinea worm has had enormous success, with only a handful of cases in four endemic countries remaining. However, two major challenges to the ongoing success of the campaign are the growing numbers of canine cases along the Tchari river in Chad and the persistence of “sporadic” human cases for which surveillance has failed to establish clear epidemiological links with previous cases. Guinea worm disease or dracunculiasis was rediscovered in Chad in 2010 after a gap of 10 years with no reported cases and in 2012 active surveillance was initiated to determine the transmission mechanics of the disease and implement interventions as part of the Guinea worm eradication program (GWEP). It was discovered that the epidemiological pattern of the disease in Chad was unlike that seen previously and a large number of dogs were infected: in the first 7 months of 2015 there werejust 7 human cases and 387 infected dogs in the country.
Thanks to colleagues at the US Centers for Disease Control (CDC) and the Carter Center, the parasite genomics group at the Wellcome Sanger Institute have produced draft genome sequences for Dracunculus medinensis and Dracunculus insignis, with D. medinensis being included as part of the 50 Helminths project and freely available at http://parasite.wormbase.org. Both genomes are still being improved.
Whole-genome sequence data from worms sampled from people and dogs in Chad and from people and other mammals from the other Guinea worm-endemic countries has already shown that, while it is possible to distinguish worms from different geographical origins there appear to be no significant differences between the worms from dogs and humans within Chad, which appears to indicate that both dog and human guinea worm cases in Chad may be derived from a single guineaworm population. This highlights the need for the GWEP to manage canine cases, but we have not definitively established transmission between canine and human cases.
To deliver results with translational impact for the Guinea Worm Eradication Program and answer key epidemiological questions we require molecular markers with sufficient power to determine direct relatedness between samples, and given the limited numbers of cases every year, nearly all biological samples need to provide high-quality analyzable sequence data: currently, only around a quarter of the samples obtained are of sufficient quality for whole genome analysis. With funding from the Carter Center we are currently working on enrichment approaches to obtain high quality genome-wide data from the majority of GW samples. We plan to sequence all GW cases reported and available to the CDC for two years, along with representative cases from previous years and from other areas, to establish baseline data for sporadic cases. Analysis of these data will focus on reconstructing the population genetic structure of the GW populations in each of the four remaining endemic countries, and reconstructing the parentage of closely related cases, aiming to establish, and if possible, quantify the levels of dog-human and human-dog transmission in Chad, establish the origins of sporadic cases and test whether a substantial bottleneck in GW cases occurred in Chad.
Contact
If you need help or have any queries, please contact us using the details below.
Sanger people
Previous Sanger people

Dr Matt Berriman
Former Senior Group Leader
External partners and funders
External
Carter Center
The Carter Center was founded in 1982 by former U.S. President Jimmy Carter and his wife, Rosalynn, in partnership with Emory University, to advance peace and health worldwide. A nongovernmental organization, the Center has helped to improve life for people in more than 80 countries by resolving conflicts; advancing democracy, human rights, and economic opportunity; preventing diseases; improving mental health care; and teaching farmers to increase crop production.
External
Centers for Disease Control and Prevention
CDC works 24/7 to protect America from health, safety and security threats, both foreign and in the U.S. Whether diseases start at home or abroad, are chronic or acute, curable or preventable, human error or deliberate attack, CDC fights disease and supports communities and citizens to do the same.
External
WHO
WHO are the directing and coordinating authority on international health within the United Nations’ system.We do this by:providing leadership on matters critical to health and engaging in partnerships where joint action is needed;shaping the research agenda and stimulating the generation, translation and dissemination of valuable knowledge;setting norms and standards and promoting and monitoring their implementation;articulating ethical and evidence-based policy options;providing technical support, catalysing change, and building sustainable institutional capacity; andmonitoring the health situation and assessing health trends.
External
Bill & Melinda Gates Foundation
To bring about the kinds of changes that will help people live healthier and more productive lives, we seek to understand the world’s inequities. Whether the challenge is low-yield crops in Africa or low graduation rates in Los Angeles, we listen and learn so we can identify pressing problems that get too little attention. Then we consider whether we can make a meaningful difference with our influence and our investments, whether it is a grant or a contract.All of our strategies—more than two dozen across the foundation—have emerged through this process of identifying what we want to accomplish for people and where we can have the greatest impact. Once we commit to an area of need, we define our major goals and identify a clear path to achieving them.