
Mathew’s expertise is in molecular cell biology, high-throughput chemical and genetic screens, cancer genomics and anti-cancer therapeutics.
Mathew joined the Sanger Institute in 2009 as a Senior Staff Scientist and was appointed a member of Faculty in 2014. Here he developed high-throughput single-agent and combination drug sensitivity screens in cancer cells to identify molecular features of cells that are predictive of drug response to help inform the development of new anti-cancer therapies. This work is on-going but has already led to the identification of new putative molecular biomarkers with potential to improve cancer treatments and contributed to the initiation of clinical trials.
In addition, he is performing CRISPR-Cas9 genetic screens in cancer cells to identify new drug targets, which led to the discovery of Werner Syndrome helicase as a synthetic-lethal target in microsatellite unstable cancers. Mathew is also passionate about developing new cancer models such as 3D organoid cultures which better capture the heterogeneity and hallmarks of patient tumours.
Mathew’s lab have developed three leading public therapeutics web portals. The Genomics of Drug Sensitivity in Cancer (GDSC) hosts drug sensitivity data and biomarker analyses to help inform the development of new cancer therapies. The Project Score database hosts CRISPR screening datasets to prioritise candidate oncology targets. The Cell Model Passports database is a hub for accessing cell model genetic and functional datasets, containing data for 1600 cell lines and most recently organoid cultures These databases and associated datasets are part of the Cancer Dependency Map, which aims to exhaustively identify vulnerabilities in human cancers to guide future precision cancer medicines
Mathew is a member of the scientific leadership team for the Open Targets, which aims to use genome-scale experiments and analysis to evaluate new therapeutic targets, as well as a member of the Cancer Research UK drug discovery small molecule expert review panel.
Prior to joining the Sanger Institute, Mathew performed his postdoctoral work in the laboratory of Professor Ashok Venkitaraman (Cambridge University, UK) with a fellowship from the Canadian Institute of Health Research. Here he developed small interfering RNA screens using high-content microscopy to understand how cells respond to anti-mitotic cancer drugs. Mathew identified the ubiquitin-conjugating enzyme UBE2S as a novel regulator of the Anaphase Promoting Complex (APC). UBE2S is necessary for ubiquitination of APC substrates, allowing progression through mitosis following mitotic checkpoint inactivation. This work identified a new component of the mitotic checkpoint machinery, provided fresh insights into the regulation of APC activity and contributed towards our understanding of the mechanisms that control the cellular response to mitotic checkpoint arrest.
Mathew completed his PhD in 2005 in the laboratory of Richard Marais at The Institute of Cancer Research (London, UK), where he was involved in the discovery and characterisation of BRAF as a human cancer gene. His PhD work elucidated how cancer-associated mutations perturb RAF activity and demonstrated that RAF-family kinases oligomerise in cells to activate the ERK-pathway, identifying a new paradigm in the regulation of RAF signalling. This was subsequently shown to have important implications for clinical deployment of therapies targeting RAF proteins in cancer.
Mathew graduated in 1999 with a BSc. in Biochemistry (Hons.) from the University of British Columbia, Canada.
Throughout Mathew’s scientific career he has strived to understand how genetic changes in cancers can be exploited to develop new cancer therapies. Several of his publications are highly cited, he has generated widely used reference datasets for the community, and his research has directly contributed to the development and testing of new cancer therapies. Mathew fosters a multi-disciplinary and collaborative approach in his research team to enable the best possible research.
My timeline
Appointed Faculty Member - Translational Cancer Genomics Laboratory
Appointed Senior Staff Scientist. Wellcome Sanger Institute.
Postdoctoral Scientist. University of Cambridge.
Doctor of Philosophy. The Institute of Cancer Research. London, UK.
Bachelor of Science, Biochemistry (Hon). University of British Columbia, Vancouver, Canada.
My publications
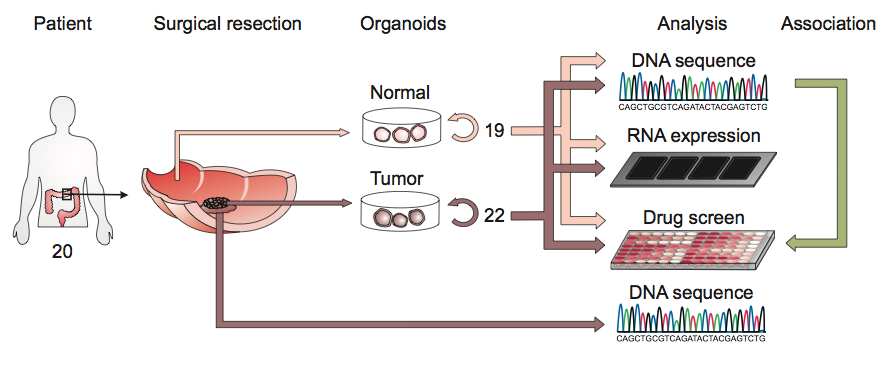
A schematic of the derivation of cancer organoids from colon patients.




