
I specialise in developing and optimising laboratory methods for genomic and metagenomic research and conducting bioinformatic and phylogenomic analyses to understand pathogen evolution and epidemiology. My diverse background includes working with viral, fungal, and bacterial human pathogens. Over the last decade, my primary research focus has been on population genomics of sexually transmitted infections (STIs), with experience working on five of the eight most common STIs, and particularly on Treponema pallidum, the causal agent of syphilis.
Sexually Transmitted Infections (STIs)
Incidence of sexually transmitted infections has been rising in many high income countries over the last two decades. My work uses whole genome sequencing to study the genetic diversity and epidemiological correlates of STI transmission. This includes projects on Neisseria gonorrhoea, Chlamydia trachomatis and Treponema pallidum. In particular, my work on T. pallidum (causative agent of syphilis) has helped to characterise the global genetic diversity and spread of this organism.
Neglected Tropical Diseases
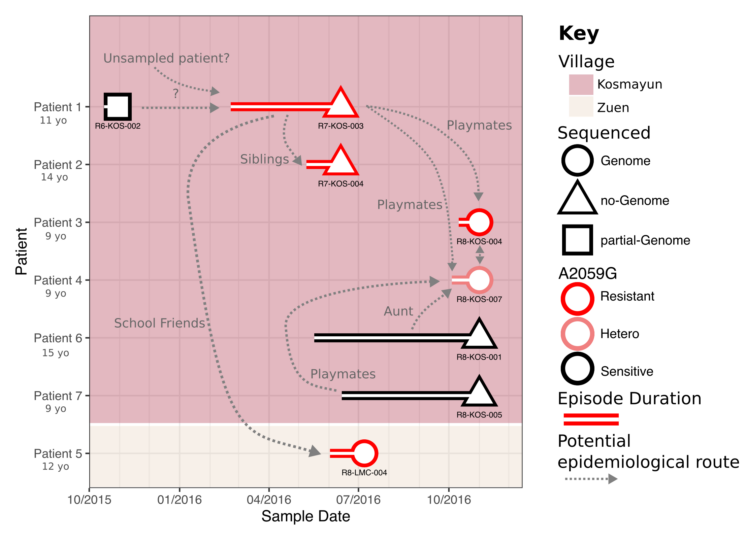
I also work on neglected tropical diseases, and infectious diseases that disproportionately affect low and middle income countries (LMICS). This includes past work on viruses (such as Hepatitis B) and fungal pathogens (such as Cryptococcus). My current work is focussed on Yaws, a form of tropical ulcer associated with Treponema pallidum subspecies pertenue. I am using genomics to explore the genetic diversity and evolution of this bacterial pathogen, with a particular emphasis on the evolution of antimicrobial resistance, the effect of erradication efforts such as mass drug administration on the bacterial population structure, and how molecular and genomic epidemiology approaches can be developed and used to inform elimination efforts.
Antimicrobial Resistance
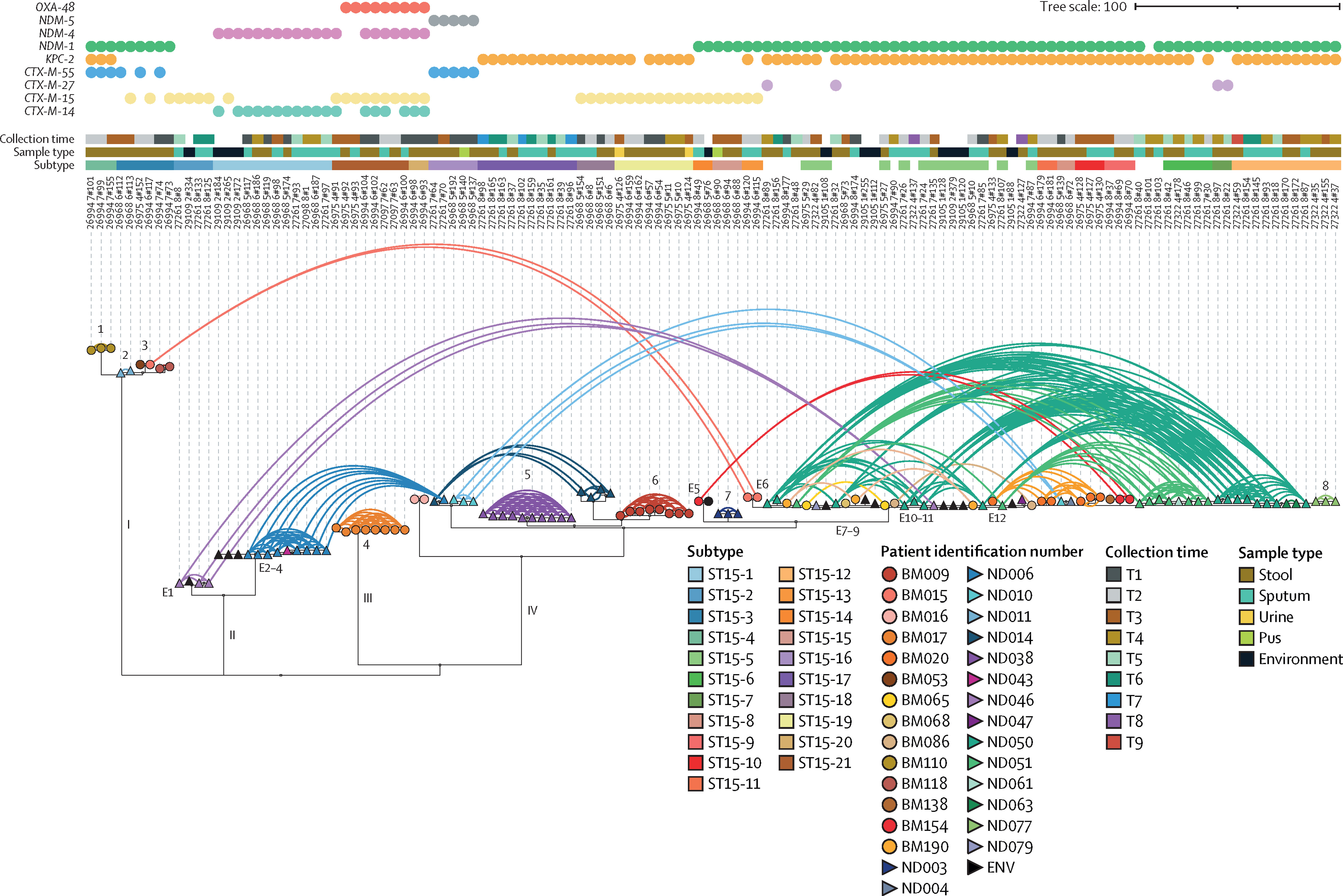
Within the Thomson group, I also support and oversee a number of projects focussing on the evolution and epidemiology of antimicrobial resistance – particularly in enterobacteriaciae such as Klebsiella pneumoniae and Escherichia coli. This includes extensive phylogenetics and analysis of mobile genetic elements. We have been investigating the acquisition of extended-spectrum Beta-lactamase resistant species in both a hospital and community environment, with a particular focus on longitudinal sampling.

My timeline
Senior Staff Scientist in Bacterial Phylogenomics and Genomic Method Development at Wellcome Sanger Institute
Postdoctoral Fellow in Bacterial Genomics at Wellcome Sanger Institute
Research Associate in Viral Genomics at University College London
Postdoctoral Research Associate in Fungal Genomics jointly between St George's University London and Imperial College
PhD Student in Virology at Public Health England and University College London
Research Assistant in Virology at Public Health England
Biomedical Scientist in Transfusion Science (inc. Transfusion Virology) at NHSBT