Yeast spotlights genetic variation’s link to drug resistance
The World Health Organisation classes antimicrobial resistance as one of the biggest threats to global health today. Currently 700,000 deaths a year worldwide*, with 25,000 of those in the EU and 23,000 in the USA are linked to antimicrobial resistance. These figures are projected** to rise to 10 million deaths a year by 2050 if nothing is done to tackle the problem. Equally, cancer is linked to 8.2 million annual deaths worldwide, with chemotherapy resistance a major limitation to treatment.
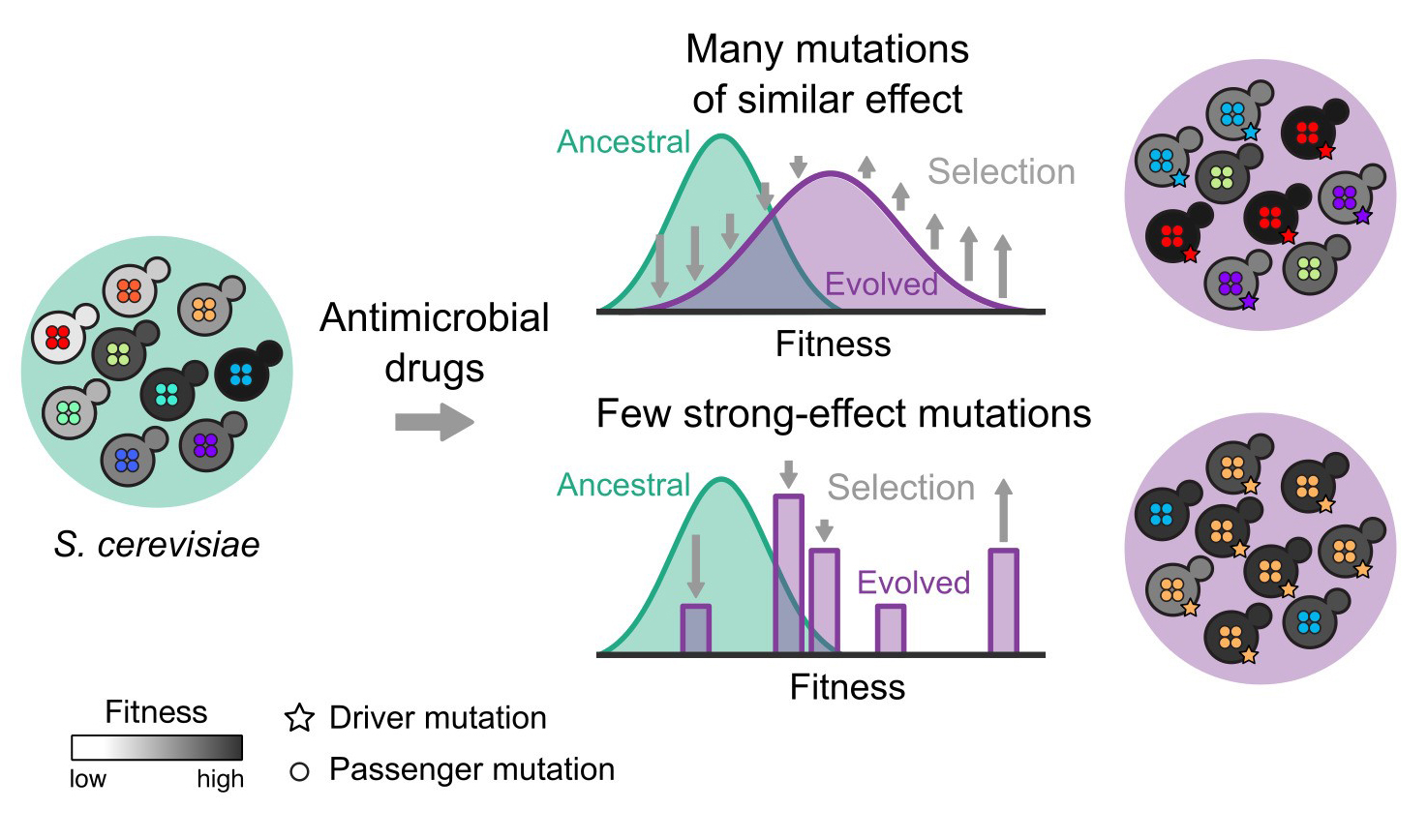
Previous studies had linked high genetic diversity within bacterial infections or in cancers with poor outcomes for patients treated with antimicrobial or chemotherapy drugs. Researchers in this study used budding yeast, creating populations of cells with more than 10 million different randomised genomes, to investigate how genetic diversity affected resistance. They evolved these to grow in antimicrobial drugs over four weeks and then studied the sensitive and resistant yeast cells.
“We found that the degree of diversity within the cell population – known as clonal heterogeneity – played a major role in the acquisition of antimicrobial resistance. By sequencing the genomes of sensitive and resistant cells we showed that some cells were pre-adapted, or primed, while other cells acquired new mutations to gain resistance.”
Dr Ignacio Vazquez-Garcia The first author from the Wellcome Trust Sanger Institute and University of Cambridge
By then crossing the evolved strains, the researchers were able to investigate the complex evolutionary processes involved in developing resistance. They were able to see not only which mutations drove resistance – called driver mutations – but also how the background mutations affected these.
They discovered two types of driver mutations. Cells with weak driver mutations needed other background mutations to grow well in antimicrobial drugs, however cells with strong driver mutations developed resistance to drugs regardless of the genetic background.
“We were able to study the evolution in time by combining genome sequences of the cell populations and tracking the growth characteristics of the yeast cells. We found that the genetic background had a major influence on whether or not weaker mutations would confer drug resistance, and in these cases many different cells adapted in a wave. However, with any genetic background, cells with strong driver mutations could “leapfrog” and outcompete other cells growing in the drugs.”
Professor Gianni Liti A senior author on the paper from the Institute for Research on Cancer and Ageing, Nice
“Our study helps understand the evolution of drug resistance, and has implications not just for yeast, but also for bacteria and cancer. Whilst further study is needed, we are building evidence to show that genetic diversity in a bacterial infection or in a tumour being treated could lay the foundation for resistance to the therapy and affect how quickly resistance develops.”
Professor Ville Mustonen Senior author from the University of Helsinki and previously at the Wellcome Trust Sanger Institute
More information
* These figures on antimicrobial resistance have been released by the CDC (https://www.cdc.gov/drugresistance/index.html) and as part of a joint report** by the UK Government and the Wellcome Trust (https://amr-review.org/).
Further figures on cancer management are available from the International Agency for Research on Cancer of the World Health Organisation (https://www.iarc.fr/en/publications/pdfs-online/wcr/2003/wcr-6.pdf).
Funding
This work was supported by the Wellcome Trust, Fundación Ibercaja, Fondation ARC, the French National Research Agency, Cancéropôle PACA, DuPont and other funding organisations. Please see the paper for the full list of funders.
Participating Centres:
- Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK
- Department of Applied Mathematics and Theoretical Physics, Centre for Mathematical Sciences, University of Cambridge, Cambridge, UK
- Université Côte d’Azur, Institute for Research on Cancer and Ageing of Nice, CNRS UMR 7284, INSERM U1081, Nice, France
- Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden
- Centre for Integrative Genetics, Department of Animal and Aquacultural Sciences, Norwegian University of Life Sciences, Ås, Norway
Publications:
Selected websites
The University of Cambridge
The mission of the University of Cambridge is to contribute to society through the pursuit of education, learning and research at the highest international levels of excellence. To date, 98 affiliates of the University have won the Nobel Prize.
Founded in 1209, the University comprises 31 autonomous Colleges, which admit undergraduates and provide small-group tuition, and 150 departments, faculties and institutions. Cambridge is a global university. Its 19,000 student body includes 3,700 international students from 120 countries. Cambridge researchers collaborate with colleagues worldwide, and the University has established larger-scale partnerships in Asia, Africa and America.
The University sits at the heart of one of the world’s largest technology clusters. The ‘Cambridge Phenomenon’ has created 1,500 hi-tech companies, 14 of them valued at over US$1 billion and two at over US$10 billion. Cambridge promotes the interface between academia and business, and has a global reputation for innovation.
Institute for Research on Cancer and Ageing, in Nice, France
IRCAN is an international institute attracting the talent, the competence and the knowhow of high level young and senior group leaders. The IRCAN’s main line of research is aimed at unravelling the pathways shared between cancer and ageing, both at the basic and translational level. The IRCAN is located at the School of Medicine of Nice and is fully supported by the University of Nice Sophia Antipolis, the INSERM (U1081), the CNRS (UMR7284), the anti-cancer center Centre Antoine Lacassagne and the Nice University Hospital.
Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate.


