Risk of cholera epidemics estimated with new rule book
Cholera has repeatedly traveled out of Asia to cause epidemics in Africa and Latin America, an international research team has found. Researchers at the Wellcome Trust Sanger Institute, Institut Pasteur in France, and collaborators from across the world, studied outbreaks in Africa, Latin America and the Caribbean from the last 60 years.
The results of two studies, published in Science (10 November), present a new ‘rule-book’ to estimate the risk of different cholera strains causing an epidemic. (Francois-Xavier Weill et al. and Daryl Domman et al.)
Despite being thought of as an ancient disease, cholera still affects 47 countries world-wide and claims the lives of nearly 100 thousand people a year*. Since the 1800s, there have been seven cholera pandemics around the globe, resulting in millions of deaths. The current and ongoing pandemic, which began in the 1960s, is caused by a single lineage of Vibrio cholerae, called 7PET.
The largest epidemic in history is ongoing in Yemen, with the total number of cases quickly approaching 1 million, and the WHO recently announcing a campaign to stop cholera by 2030**.
In two new studies, scientists analysed historical cholera samples from across the globe. The team studied genetic data from over 1200 cholera samples, some dating back to the 1950s.
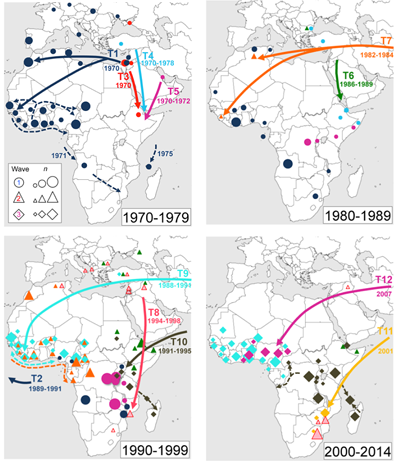
The researchers focused on Africa and Latin America, due to the large epidemics that have occurred in those regions. The seventh pandemic of cholera first came to Africa in 1970, and Africa has since become the continent most affected by the disease. Cholera appeared in Latin America in 1991, after an absence of 100 years.
The team found that 7PET strains from Asia were repeatedly introduced into two main regions of Africa: West Africa and East/Southern Africa.
“Our results show that multiple new versions of 7PET bacteria have entered Africa since the 1970s. Once introduced, cholera outbreaks follow similar paths when spreading across that continent. The results give us a sense of where we can target specific regions of Africa for improved surveillance and control.”
François-Xavier Weill First author on the African study and principal investigator from the Institut Pasteur
Scientists also investigated the evolution of antibiotic resistance in cholera. The team found that all recent introductions of cholera bacteria into Africa were already resistant to antibiotics. Using the genomic data they were able to track the source of the antibiotic resistance back to South Asia.
In Latin America, the team not only focused on the 7PET strains that cause epidemics, but other strains of Vibrio cholerae that cause sporadic low level disease. This allowed the researchers to uncover that different strains of Vibrio cholerae can be assigned different risks for causing large outbreaks. This has significant implications for cholera control efforts world-wide.
“Our data show that when a 7PET pandemic strain enters into Latin America from elsewhere, it can cause massive epidemics, like those seen in Peru in the 1990s and Haiti in 2010. However, we now know that other strains already in this region can still make people sick, but seem to not have this epidemic potential. Knowing which strain is which allows for an appropriate public health response from regional or national governments.”
Daryl Domman First author on the Latin American study from the Wellcome Trust Sanger Institute
“These findings have implications for the control of cholera pandemics. We are now getting a real sense of how cholera is moving across the globe, and with that information we can inform improved control strategies as well as basic science to better understand how a simple bacterium continues to pose such a threat to human health.”
Professor Nick Thomson Senior author from the Wellcome Trust Sanger Institute
More information
About Cholera:
Cholera is an acute diarrhoeal disease caused by ingesting food or water contaminated with the bacterium, Vibrio cholerae. Some people develop water diarrhoea with severe dehydration, which can lead to death if left untreated. The disease spreads between humans in areas with inadequate access to clean water and sanitation. https://www.who.int/health-topics/cholera
* https://www.who.int/en/news-room/feature-stories/detail/prevention-for-a-cholera-free-world
Publications:
Funding:
Study 1 – Institut Pasteur and the Institut Pasteur International Network, the Institut de Veille Sanitaire, French government’s Investissement d’Avenir programme (Laboratoire d’Excellence), the Fondation Le Roch-Les Mousquetaires, Wellcome (grant 098051), Indian Council of Medical Research
Study 2 – Wellcome (grant 098051), Institut Pasteur, Santé Publique France, French government’s Investissement d’Avenir programme (Laboratoire d’Excellence)
Publications:
Selected websites
About the Institut Pasteur and the Institut Pasteur International Network
The Institut Pasteur, a private foundation with officially recognized charitable status set up by Louis Pasteur in 1887, is today an internationally renowned center for biomedical research with a network of 33 institutes worldwide. In the pursuit of its mission to prevent and fight against diseases in France and throughout the world, the Institut Pasteur operates in four main areas: scientific and medical research, public health and health monitoring, teaching, and business development and technology transfer. More than 2,500 people work on its Paris campus. The Institut Pasteur is a globally recognized leader in infectious diseases, microbiology, and immunology. Its 130 units also focus their research on certain cancers, genetic and neurodegenerative diseases, genomics and developmental biology. This research aims to expand our knowledge of living organisms in a bid to lay the foundation for new prevention strategies and novel therapeutics. Since its inception, 10 Institut Pasteur scientists have been awarded the Nobel Prize for Medicine, including two in 2008 for the 1983 discovery of the human immunodeficiency virus (HIV) that causes AIDS. www.pasteur.fr/en
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease. www.sanger.ac.uk
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate. wellcome.org


