Cancer cell lines predict drug response
Led by scientists from the Wellcome Trust Sanger Institute, the European Bioinformatics Institute (EMBL-EBI) and the Netherlands Cancer Institute, the international study discovered a strong link between many mutations in patient cancer samples, and the sensitivity to particular drugs. This could advance personalised cancer medicine by leading to results that help doctors predict the best available drugs, or the most suitable clinical trials for each individual patient.
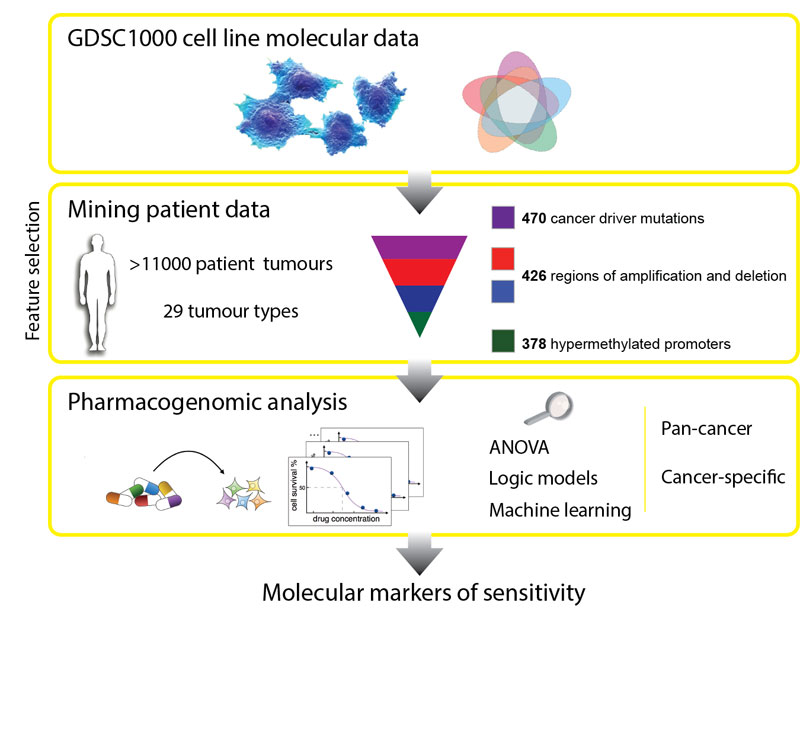
In the first systematic, large-scale study to combine molecular data from patients, laboratory cancer cell lines and drug sensitivity, the researchers looked at genetic mutations known to cause cancer in more than 11,000 patient samples of 29 different tumour types.
They built a catalogue of the genetic changes that cause cancer in patients and mapped these alterations onto 1000 cancer cell lines. Next, they tested the cell lines for sensitivity to 265 different cancer drugs to understand which of these changes effect sensitivity.
The researchers made two significant discoveries. Firstly, that the majority of molecular abnormalities found in patient’s cancers are also found in cancer cells in the laboratory. This means that cell lines are indeed useful models to identify which drugs would work best for patients. Secondly, many of the molecular abnormalities detected in the thousands of patient cancer samples can, both individually but also in combination, have a strong effect on whether a particular drug affects a cancer cell’s survival.
The results suggest cancer cell lines could be better exploited to learn which drugs offer the most effective treatment to which patients.
“In this study we compared the genetic landscape of patient tumours with that of cancer cells grown in the lab. We found that cell lines do carry the same genetic alterations that drive cancer in patients. This means that drug sensitivity testing in cell lines can be used to figure out how a tumour is likely to respond to a drug.”
Dr Mathew Garnett Joint leader of the study from the Wellcome Trust Sanger Institute

“If a cell line has the same genetic features as a patient’s tumour, and that cell line responded to a specific drug, we can focus new research on this finding. This could ultimately help assign cancer patients into more precise groups based on how likely they are to respond to therapy. This resource can really help cancer research. Most importantly, it can be used to create tools for doctors to select a clinical trial which is most promising for their cancer patient. That is still a way off, but we are heading in the right direction.”
Dr Francesco Iorio Joint first author and postdoctoral researcher at both EMBL-EBI and the Sanger Institute
“We need better ways to figure out which groups of patients are more likely to respond to a new drug before we run complex and expensive clinical trials. Our research shows that cancer cell lines do capture the molecular alterations found in tumours, and so can be predictive of how a tumour will respond to a drug. This means the cell lines could tell us much more about how a tumour is likely to respond to a new drug before we try to test it in patients. We hope this information will ultimately help in the design of clinical trials that target those patients with the greatest likelihood of benefiting from treatment.”
Dr Ultan McDermott Joint leader of the study and clinician scientist from the Sanger Institute
More information
Further information
The study was based on publicly available data gathered over the past six years by global scientific collaborations: the Cancer Genome Atlas and the International Cancer Genome Consortium. Data from the study are freely available via the Wellcome Trust Sanger Institute and European Bioinformatics Institute (EMBL-EBI).
To enable other scientists to explore this resource, all data and appropriate tools are released via a website (http://www.cancerrxgene.org) and as Python tool (GDSCTools; http://gdsctools.readthedocs.io/).
The EBI-Sanger Postdoctoral (ESPOD) Programme offers joint projects at the EBI and Sanger Institute that combine experimental (wet-lab) and computational (dry-lab) approaches. It builds on the collaborative relationship between the two institutes on the Wellcome Genome Campus.
Funding
Funding for this project was provided by the Wellcome Trust (Grant: 086375 and 102696), the European Molecular Biology Laboratory, National Cancer Institute (U24CA143835) , the Netherlands Organization for Scientific Research, the Seventh Framework Programme of the European Union (FP7/2007-2013) , La Fundació la Marató de TV3, European Research Council, the Ministerio de Ciencia e Innovacion grant number SAF2011-22803, the Institute of Health Carlos III (ISCIII), the Spanish Cancer Research Network (RTICC), the Health and Science Departments of the Catalan Government Generalitat de Catalunya, the Cellex Foundation and Cancer Research UK.
Publications:
Selected websites
European Molecular Biology Laboratory
EMBL is Europe’s flagship laboratory for the life sciences, with more than 80 independent groups covering the spectrum of molecular biology. EMBL is international, innovative and interdisciplinary – its 1800 employees, from many nations, operate across five sites: the main laboratory in Heidelberg, and outstations in Grenoble; Hamburg; Hinxton, near Cambridge (the European Bioinformatics Institute), and Monterotondo, near Rome. Founded in 1974, EMBL is an inter-governmental organisation funded by public research monies from its member states. The cornerstones of EMBL’s mission are: to perform basic research in molecular biology; to train scientists, students and visitors at all levels; to offer vital services to scientists in the member states; to develop new instruments and methods in the life sciences and actively engage in technology transfer activities, and to integrate European life science research. Around 200 students are enrolled in EMBL’s International PhD programme. Additionally, the Laboratory offers a platform for dialogue with the general public through various science communication activities such as lecture series, visitor programmes and the dissemination of scientific achievements.
EMBL-EBI
The European Bioinformatics Institute is part of EMBL, and is a global leader in the storage, analysis and dissemination of large biological datasets. EMBL-EBI helps scientists realise the potential of ‘big data’ by enhancing their ability to exploit complex information to make discoveries that benefit mankind. We are a non-profit, intergovernmental organisation funded by EMBL’s 21 member states and two associate member states. Our 570 staff hail from 57 countries, and we welcome a regular stream of visiting scientists throughout the year. We are located on the Wellcome Genome Campus in Hinxton, Cambridge in the United Kingdom.
The Netherlands Cancer Institute, Amsterdam
The Netherlands Cancer Institute is at the international forefront of cancer care and research for already more than a century. The unique combination of health care and scientific research within the same institute offers great benefit for cancer patients. Specialized cancer care professionals work together in multidisciplinary teams every day to set up and carry out treatment plans tailored to the needs of individual patients because no two tumors are alike. Cancer patients or people suspected of having cancer can come to our hospital, known as the Antoni van Leeuwenhoek, to make use of this personal approach and the state-of-the-art research and treatment facilities. The research institute employs more than 560 scientists investigating many aspects of cancer development, diagnosis, treatment and epidemiology. Scientists at the Netherlands Cancer Institute have excess to state-of-the-art research facilities supporting their basic, translational and clinical research. This scientific research could not be carried out without the institutional support of the Dutch Cancer Society, the Ministry of Health, Welfare and Sport, the many research grants obtained by our researchers from (inter)national funding agencies, and the generous donations made by individuals that support our research program. The Netherlands Cancer Institute is the only OECI designated Comprehensive Cancer Center in the Netherlands.
The Wellcome Trust Sanger Institute
The Wellcome Trust Sanger Institute is one of the world’s leading genome centres. Through its ability to conduct research at scale, it is able to engage in bold and long-term exploratory projects that are designed to influence and empower medical science globally. Institute research findings, generated through its own research programmes and through its leading role in international consortia, are being used to develop new diagnostics and treatments for human disease.
The Wellcome Trust
The Wellcome Trust is a global charitable foundation dedicated to achieving extraordinary improvements in human and animal health. We support the brightest minds in biomedical research and the medical humanities. Our breadth of support includes public engagement, education and the application of research to improve health. We are independent of both political and commercial interests.


