Hospitals key in the spread of extremely drug resistant bacteria in Europe
New research has found that antibiotic-resistant strains of Klebsiella pneumoniae, an opportunistic pathogen that can cause respiratory and bloodstream infections in humans, are spreading through hospitals in Europe. Certain strains of K. pneumoniae are resistant to the carbapenem antibiotics that represent the last line of defence in treating infections and are therefore regarded as extremely drug resistant (XDR).
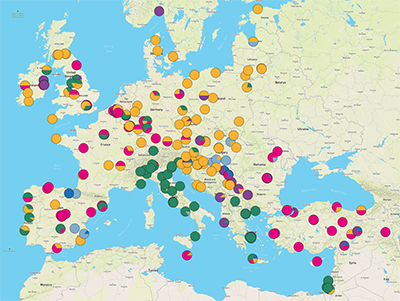
During a Europe-wide survey of the Enterobacteriaceae family of bacteria, researchers at the Centre for Genomic Pathogen Surveillance, based at the Wellcome Sanger Institute, University of Freiburg and their partners, analysed the genomes of almost 2,000 K. pneumoniae samples taken from patients in 244 hospitals in 32 countries. The results, published today (29 July 2019) in Nature Microbiology, will inform public health efforts to control the spread of these infections in hospitals across Europe.
It is estimated that 341 deaths in Europe were caused by carbapenem-resistant K. pneumoniae in 2007; by 2015 the number of deaths had increased six-fold to 2,094*. The high number of deaths is down to the fact that once carbapenems are no longer effective against antibiotic-resistant bacteria, there are few other options left. Infants, the elderly and immuno-compromised individuals are particularly at risk.
Researchers identified a small number of genes that, when expressed, can cause resistance to carbapenem antibiotics. These genes produce enzymes called carbapenemases, which ‘chew up’ the antibiotics, rendering them useless.
Of concern to public health is the recent emergence of a small number of ‘high-risk’ clones carrying one or more carbapenemase genes, which have spread rapidly. It is thought that the heavy use of antibiotics in hospitals favours the spread of these highly-resistant bacteria, which outcompete other strains that are more easily treatable with antibiotics.
“The ‘One Health’*** approach to antibiotic resistance focuses on the spread of pathogens through humans, animals and the environment, including hospitals. But in the case of carbapenem-resistant Klebsiella pneumoniae, our findings imply hospitals are the key facilitator of transmission – over half of the samples carrying a carbapenemase gene were closely related to others collected from the same hospital, suggesting that the bacteria are spreading from person-to-person primarily within hospitals.”
Dr Sophia David First author of the study, based at the Centre for Genomic Pathogen Surveillance
Antibiotic-resistant bacteria samples were also much more likely to be closely related to samples from a different hospital in the same country rather than across countries – suggesting that national healthcare systems as a whole play an important role in the spread of these antibiotic-resistant bacteria.
Despite the clear threat that carbapenem-resistant K. pneumoniae pose to patients, more effective infection control in hospitals, including consideration of how patients move between hospitals and hygiene interventions, will have an impact.
“We are optimistic that with good hospital hygiene, which includes early identification and isolation of patients carrying these bacteria, we can not only delay the spread of these pathogens, but also successfully control them. This research emphasises the importance of infection control and ongoing genomic surveillance of antibiotic-resistant bacteria to ensure we detect new resistant strains early and act to combat the spread of antibiotic resistance.”
Professor Hajo Grundmann Co-lead author and Head of the Institute for Infection Prevention and Hospital Hygiene at the Medical Centre, University of Freiburg
The second survey of the Enterobacteriaceae bacteria family across hospitals in Europe is currently being planned. The data generated is made available through MicroReact****, a publicly-available, web-based tool developed by the Centre for Genomic Pathogen Surveillance. MicroReact will help researchers and healthcare systems to chart the spread of antibiotic resistance in pathogens like K. pneumoniae and monitor how they are evolving.
“Genomic surveillance will be key to tackling the new breeds of antibiotic-resistant pathogen strains that this study has identified. Currently, new strains are evolving almost as fast as we can sequence them. The goal to establish a robust network of genome sequencing hubs will allow healthcare systems to much more quickly track the spread of these bacteria and how they’re evolving.”
Professor David Aanensen Co-lead author and Director of the Centre for Genomic Pathogen Surveillance
More information
Notes to Editors:
*Statistics on deaths caused by carbapenem-resistant K. pneumoniae taken from Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56-66 (2019). https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(18)30605-4/fulltext
**The survey is titled EuSCAPE (European Survey of Carbapenemase-Producing Enterobacteriaceae).
***For information on the One Health initiative see http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/about-amr/one-health
****The data is freely available via MicroReact visualisation platform: https://www.sanger.ac.uk/science/tools/microreact
Publication:
Sophia David et al. (2019) Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nature Microbiology. DOI: 10.1038/s41564-019-0492-8
Funding:
This work was funded by The Centre for Genomic Pathogen Surveillance, Wellcome Genome Campus, Wellcome (grants 098051 and 099202) and The NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance (NIHR 16/136/111). The EuSCAPE project was funded by ECDC through a specific framework contract (ECDC/2012/055) following an open call for tender (OJ/25/04/2012-PROC/2012/036).
Selected websites
Centre for Genomic Pathogen Surveillance (CGPS)
To tackle anti-microbial resistance (AMR), we provide universal surveillance strategies at scale, facilitating rapid analysis and enabling the training of experts worldwide. The CGPS is committed to making our expertise in genomic pathogen surveillance as accessible as our data and tools. Find out more at https://www.pathogensurveillance.net/
Institute for Infection Prevention and Hospital Epidemiology, University of Freiburg Medical Centre
The Institute for Infection Prevention and Hospital Epidemiology of the University of Freiburg Medical Centre is one of Europe’s leading centres devoted to large scale surveys on the epidemiology of antibiotic-resistant bacteria. It develops crucial tools for the analysis, prediction and containment of continental spread of hospital infections across Europe. Find out more at https://www.uniklinik-freiburg.de/iuk-en.html
The Wellcome Sanger Institute
The Wellcome Sanger Institute is a world leading genomics research centre. We undertake large-scale research that forms the foundations of knowledge in biology and medicine. We are open and collaborative; our data, results, tools and technologies are shared across the globe to advance science. Our ambition is vast – we take on projects that are not possible anywhere else. We use the power of genome sequencing to understand and harness the information in DNA. Funded by Wellcome, we have the freedom and support to push the boundaries of genomics. Our findings are used to improve health and to understand life on Earth. Find out more at www.sanger.ac.uk or follow us on Twitter, Facebook, LinkedIn and on our Blog.
Wellcome
Wellcome exists to improve health for everyone by helping great ideas to thrive. We’re a global charitable foundation, both politically and financially independent. We support scientists and researchers, take on big problems, fuel imaginations and spark debate. wellcome.org



