
Zebrafish Mutation Project
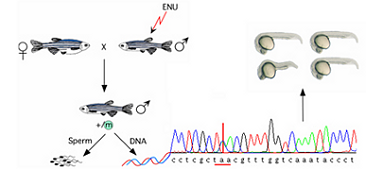 With knockout alleles in more than 60% of protein-coding genes, this project is uniquely capable of annotating gene function to thousands of vertebrate genes of which we currently know very little.
With knockout alleles in more than 60% of protein-coding genes, this project is uniquely capable of annotating gene function to thousands of vertebrate genes of which we currently know very little.
Morphological Phenotyping
As a first pass analysis to annotate gene function to the ZMP allele collection, a high throughput phenotyping pipeline has been established to evaluate developmental morphology in the first 5 days of development. With its rapid and translucent development, by day 5 a zebrafish embryo has developed the majority of vertebrate structures and organs; eyes, brain, heart, liver, kidney, pancreas, blood cells, etc… Morphological features of the developing larvae are assessed and changes in their features are linked back to identified mutations.
Worldwide Collaboration
An important aspect of the ZMP is to make our vast collection of alleles available to researchers across the entire world. This is achieved through our partners, the Zebrafish International Resource Center (ZIRC) and the European Zebrafish Resource Center (EZRC), who provide distribution services for the zebrafish community.
Contact
If you need help or have any queries, please contact us using the details below.
External partners and funders
External
Wellcome Sanger Institute
External
National Institutes of Health
External
Freek van Eeden
External
Cuppen Group
Affiliated Sites
External
Knockout List
External
Search
External
Register
External
Order Allele
External